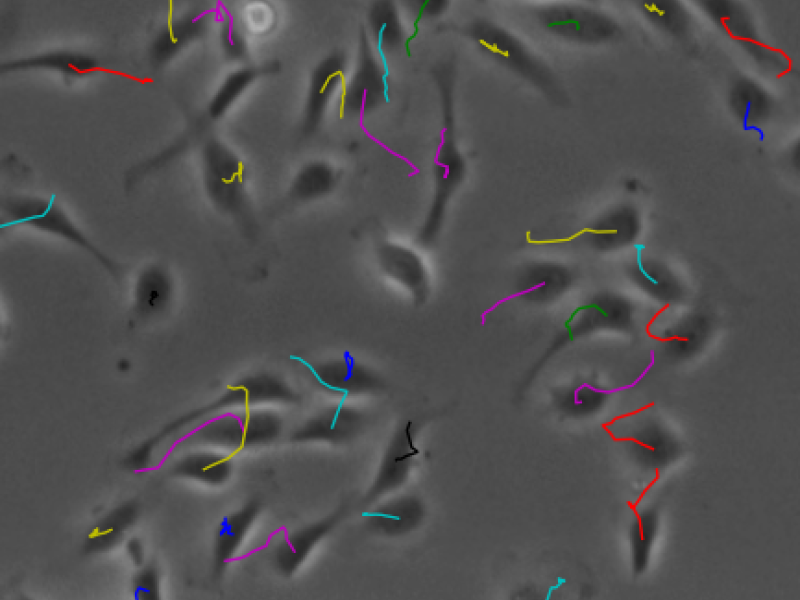
In biological research, microscopy is a powerful tool to investigate morphological changes in tissues and cell cultures. Modern setups are able to produce 100000 and more images per day with increasing image quality. The images need to be stored, annotated and analyzed. This massive amount of microscopy data is too large to be analyzed manually. There is a clear need for automatic software-based analysis. This project aims to provide a collaborative platform for scientists to work with large collections of microscopy images and assist them with state-of-the-art analysis algorithms. While research questions are evolving, the analyses need to evolve as well. Whereas early applications of microscopy image analysis were simple cell counts, today phenotypes are expected to be detected automatically with machine learning approaches. We hope to achieve best results by implementing a collaborative platform on which biologists and computer scientists work together. It will provide a full pipeline from intuitive data management to the analysis and computation of large data sets. Managing many large data sets together will also enable us to gather meta data and perform more powerful analysis in a bigger context. Collaboration will not only happen between biologists and computer scientists. Data can also be shared between biologists or even published, for example, to provide supplemental data to a journal publication. The developers of the analyses can furthermore collaborate on code and manage the code for the analysis modules.