Where Quantum Physics Meets Photosynthesis
Recent experimental results suggest that quantum-mechanical coherence plays a role in efficient energy transfer in photoactive complexes. The physical simulation of energy pathways requires designing new scalable algorithms for current and future many-core supercomputers. ZIB tackles this challenge in interdisciplinary projects bridging physics, distributed scientific computing, and visual analysis.
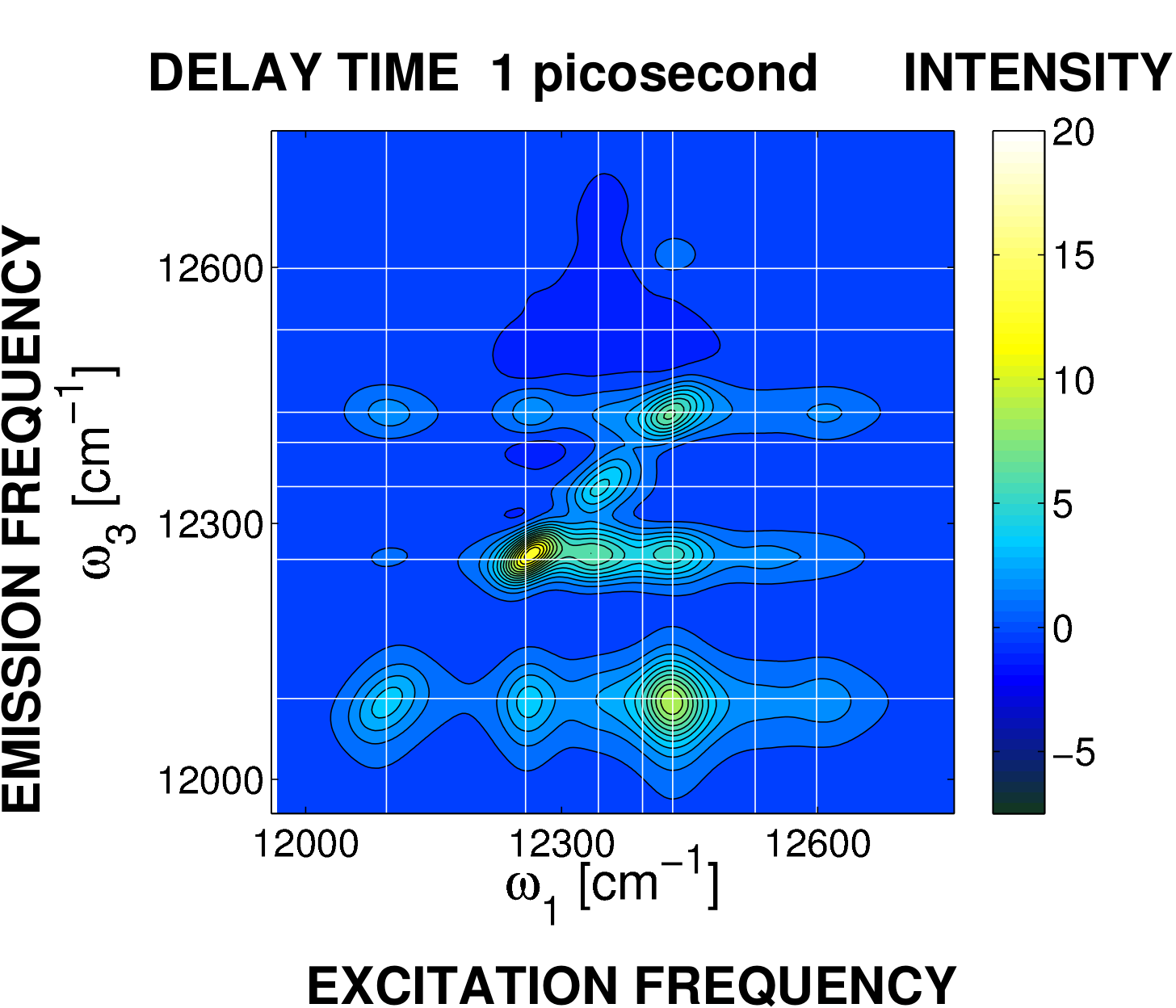
[1] C. Kreisbeck, T. Kramer, M. Rodríguez, B. Hein. High-performance solution of hierarchical equations of motions for studying energy-transfer in light-harvesting complexes. Journal of Chemical Theory and Computation. 7: 2166, 2011.F1: ime-resolved spectroscopy of the Fenna-Matthews Olson complex computed with GPU-HEOM [1]. After one picosecond, intensity peaks below the diagonal of the two-dimensional frequency representation show the conversion of excitation energy into molecular vibrations.
Photosynthesis fuels life on Earth by converting incoming solar radiation into chemical energy. Different strategies emerged during the evolution of natural photosynthesis to collect light in antenna systems and to guide it to the reaction center. Recent pulsed-laser experiments on parts of photosynthetic complexes reveal a time-resolved picture of the energy transport.
By tracking the frequency difference between absorbed and re-emitted light, the partial conversion of the excitation energy into heat has been demonstrated.
The heat generation guides the energy transfer to the reaction center, which lies energetically lower [Figure 1]. Unexpectedly, on top of the irreversible heating process, indications of intermolecular, quantum-mechanical coherences have been observed.
[2] C. Kreisbeck, T. Kramer: Long-Lived Electronic Coherence in Dissipative Exciton-Dynamics of Light-Harvesting Complexes. Journal of Physical Chemistry Letters 3: 2828, 2012.The presence of coherence suggests that classical transport models need to be revised to take into account quantummechanical effects that do assist the energy transfer [2]. The better understanding of quantum effects on the transfer through molecular networks at ambient room temperature will help to optimize OLEDs and organic photovoltaics.
Finding energy pathways in photosystems
The major obstacle for analyzing the role of coherence in photosynthetic energy transport is the computational difficulty of obtaining an accurate numerical solution for the underlying quantum-mechanical dynamics.
[3] D. E. Tronrud, J. Wen, L. Gay, R. E. Blankenship: The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth. Res. 100: 79-87, 2009.Fully resolved quantum-dynamical models for complexes comprising hundreds of excited molecules are not feasible, but for smaller molecular units found in the Fenna-Matthews-Olson complex of green sulfur bacteria [3], computations of optical spectra have been performed [4].
[5] T. Kramer, C. Kreisbeck. Modelling excitonic-energy transfer in light-harvesting complexes. AIP Conference Proceedings. 1575:111-135, 2014.In a coarse-grained description, the ensemble of quantum-mechanical states of the chlorophyll molecules is captured by the corresponding density matrix, which generalizes the concept of a wave function [5]. The forward time evolution of the density matrix is governed by the Liouville-von Neumann equation [Figures 2 + 3].

Figure 2: The quantum-mechanical time evolution of a density matrix is given by a hierarchy of Liouville-von Neumann equations.
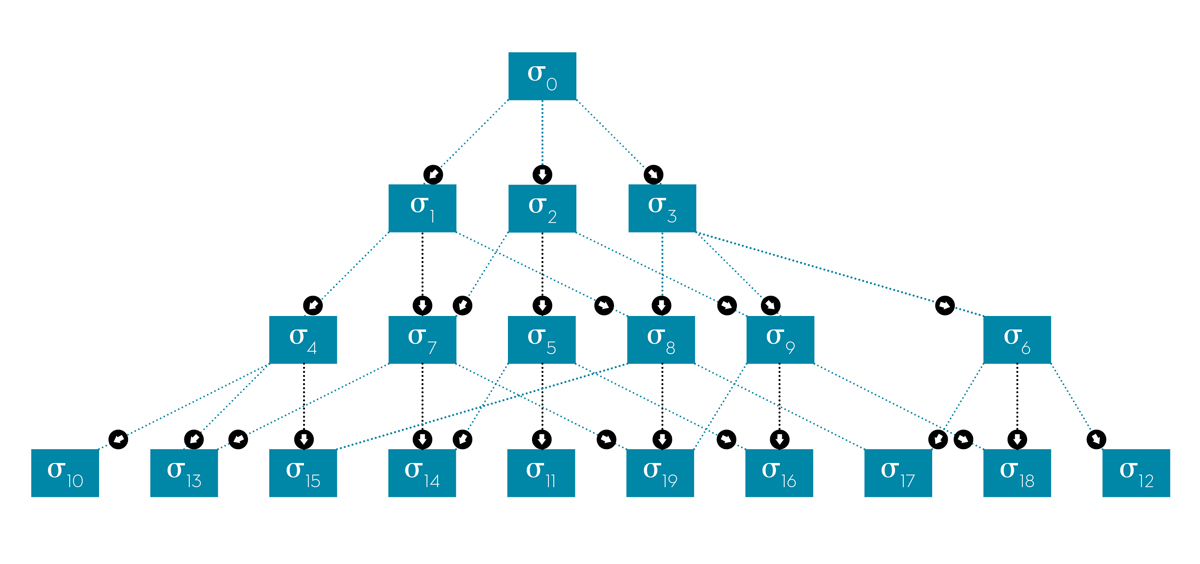
Figure 3: The coupling of the different hierarchy elements is represented by a connectivity graph.
[6] C. Kreisbeck, T. Kramer. Exciton Dynamics Lab for Light-Harvesting Complexes [GPU-HEOM]. https://nanohub.org/resources/gpuheompop, 2013.Within the joint computer science/physics projects at ZIB, funded by the German Research Foundation (DFG), we design and apply distributed algorithms for calculating the excitonic energy flow in photoactive systems on massively parallel many-core computing platforms [Figure 4]. To obtain a solution for the Liouville-von Neumann equation, we have adapted the Hierarchical Equations Of Motion (HEOM) method introduced by Kubo and Tanimura to work on general purpose graphics processing units (GPGPUs). A demonstration tool for computing the excitation dynamics is installed as a ready-to-run cloud computing application on the open access nanoHUB.org simulation platform and has attracted a worldwide user base [6].
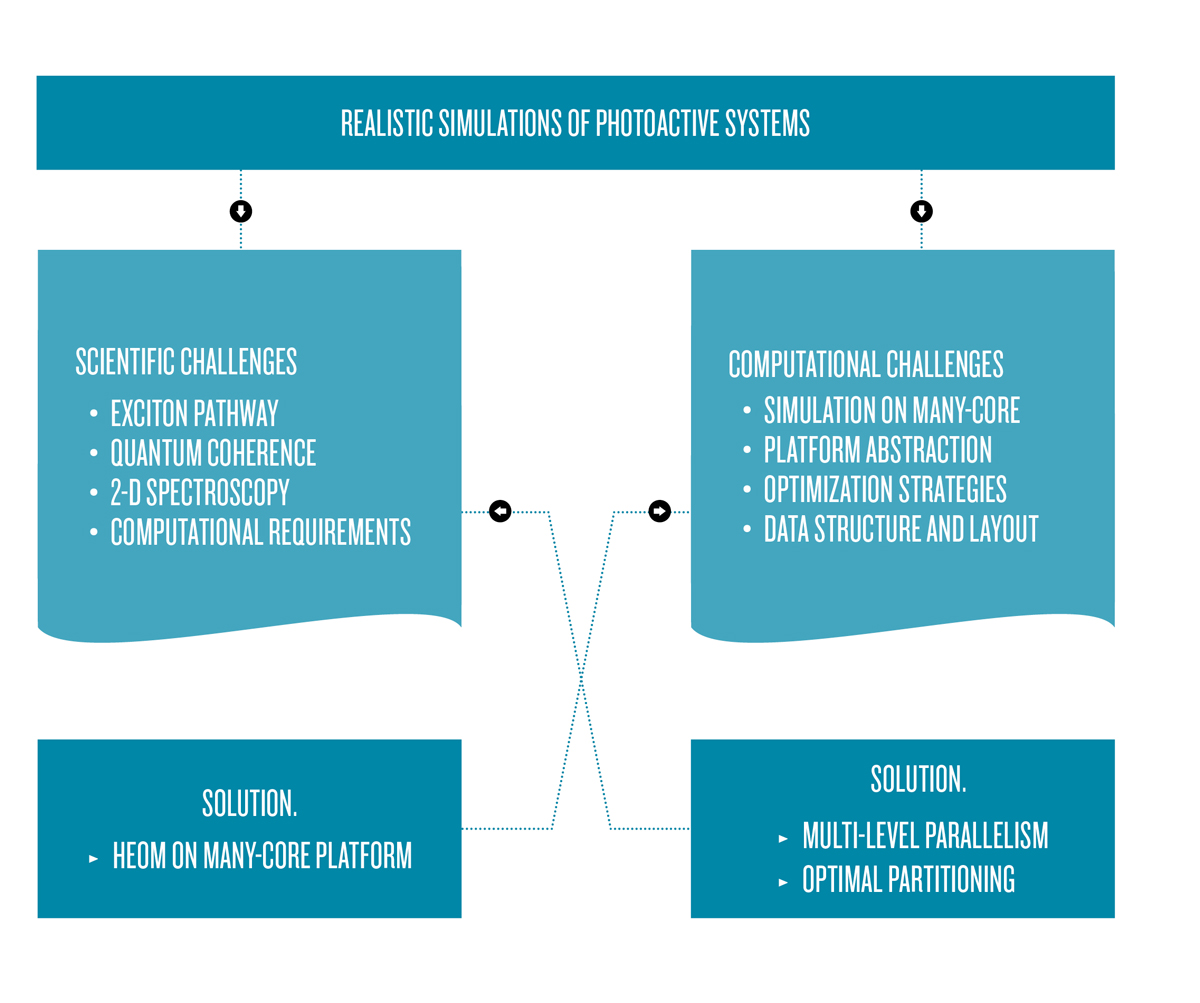
Figure 4: Realistic simulations of energy pathways through photoactive complexes require the design of new algorithms suitable for the variety of parallel computing devices used in today’s and tomorrow’s supercomputers.
Algorithm design for next generation HPC systems with many-cores
Current state-of-the-art is the simulation of the optical response of the 7-8 bacteriochlorophyll network found in the Fenna-Matthews Olson complex (FMO). Larger systems exceed the capacity of a compute node with a single accelerator device like a GPGPU or many-core accelerator (e.g. Intel Xeon Phi). The incorporation of the molecular vibrations leads to high memory consumption and requires developing methods which scale across multiple accelerator devices and compute nodes. Figure 5 illustrates the increasing memory requirements for larger molecular complex sizes.
The increasing use of coprocessors in high-performance computers, as shown in the TOP 500 list, for example, and efforts by the leading vendors (Intel, Nvidia, AMD) to fuse multi-core CPUs and coprocessors into a single chip, both signal the transition from multi-core to many-core processors. Along with this development, existing optimization strategies and algorithmic designs need to be adapted to take advantage of the various levels of parallelism provided by the new architectures:
- Parallelism across hundreds to thousands of many-core processors;
- Parallelism within many-core processors; and
- Parallelism at the level of short-vector SIMD (single-instruction multiple-data) cores.
In addition, different characteristics of the architectures have to be considered, e.g. number and performance of processing elements, memory hierarchies and bandwidths, intranode and internode communication, and programming models. Due to the variety of hardware platforms, an important research goal is to find sweet spots between platform-specific optimizations and generic massively-parallel algorithms.
[7] C. Kreisbeck, T. Kramer, A. Aspuru-Guzik. Scalable high-performance algorithm for the simulation of exciton-dynamics. Application to the light harvesting complex II in the presence of resonant vibrational modes. Journal of Chemical Theory and Computation. 10:4045-4054, 2014.In this project, we take the GPGPU implementation of the HEOM density-matrix evolution as a baseline for researching various optimization strategies for various many-core platforms [7]. In addition, we evaluate current many-core programming models with respect to usability, portability, and performance. One immediate goal is to generalize the HEOM implementation to support GPGPUs, Intel Xeon Phi many-core and multi-core CPUs – across multiple compute nodes.
The program development across different platforms, along with the claim of retaining a similar program flow, can be addressed with the OpenCL programming framework, for example, which provides an open standard for cross-platform parallel programming within heterogeneous compute nodes. OpenCL aims at implementing the kernels just once, and to compile them at runtime for the particular platform detected in the computer system. This cross-platform portability is compromised by the performance guidelines of the device vendors, whose optimization recommendations are sometimes orthogonal. Even for the same hardware platform, OpenCL optimization strategies and techniques can be very different
![Molecular complex size vs. memory requirements for the HEOM method. Solutions for larger photosynthetic complexes found in the light harvesting system 2 [9] and photosystem II [10] can only be acquired by scaling up to many compute nodes. Molecular complex size vs. memory requirements for the HEOM method. Solutions for larger photosynthetic complexes found in the light harvesting system 2 [9] and photosystem II [10] can only be acquired by scaling up to many compute nodes.](/sites/default/files/feature_attachments/HEADING4THESUN/Heading-for-the-Sun-5.jpg)
[8] V. Cherezov, J. Clogston, M. Z. Papiz, M. Caffrey. Room to Move: Crystallizing Membrane Proteins in Swollen Lipidic Mesophases. J.Mol.Biol. 357:1605-1618, 2006.Figure 5: Molecular complex size vs. memory requirements for the HEOM method. Solutions for larger photosynthetic complexes found in the light harvesting system 2 [8] and photosystem II [9] can only be acquired by scaling up to many compute nodes.
The limitations of cross-device performance demonstrate the need for additional close-to-hardware optimization, which requires the rewriting of relevant code sections for a particular hardware. A case study [10] for parts of the HEOM algorithm covering the coherent dynamics shows that different optimization techniques for every target hardware must be selected to achieve optimal performance. Figure 6 shows the performance portability of OpenCL across different computing devices.
Additional complexity arises from extending a parallel application across multiple compute nodes (possibly using PGAS or MPI). Future many-core systems require identifying generic optimization strategies which work across various many-core platforms and heterogeneous combinations thereof. Since direct porting of existing algorithms and workflows to these future architectures is not enough, the combined expertise and effort of computer science and user groups — biophysics and theoretical chemistry — is necessary to identify theoretical approaches with inherent parallelism and scalability.
We plan to develop configurable data layout templates that take the needed optimizations across all scales of massively parallel many-core HPC systems into account — from SIMD parallelism to distributed memory partitioning, based on connectivity graphs. Other contributions are theoretical upper and lower bounds for the communication costs of HEOM. The results are expected to be general enough to be applicable to other problem domains.
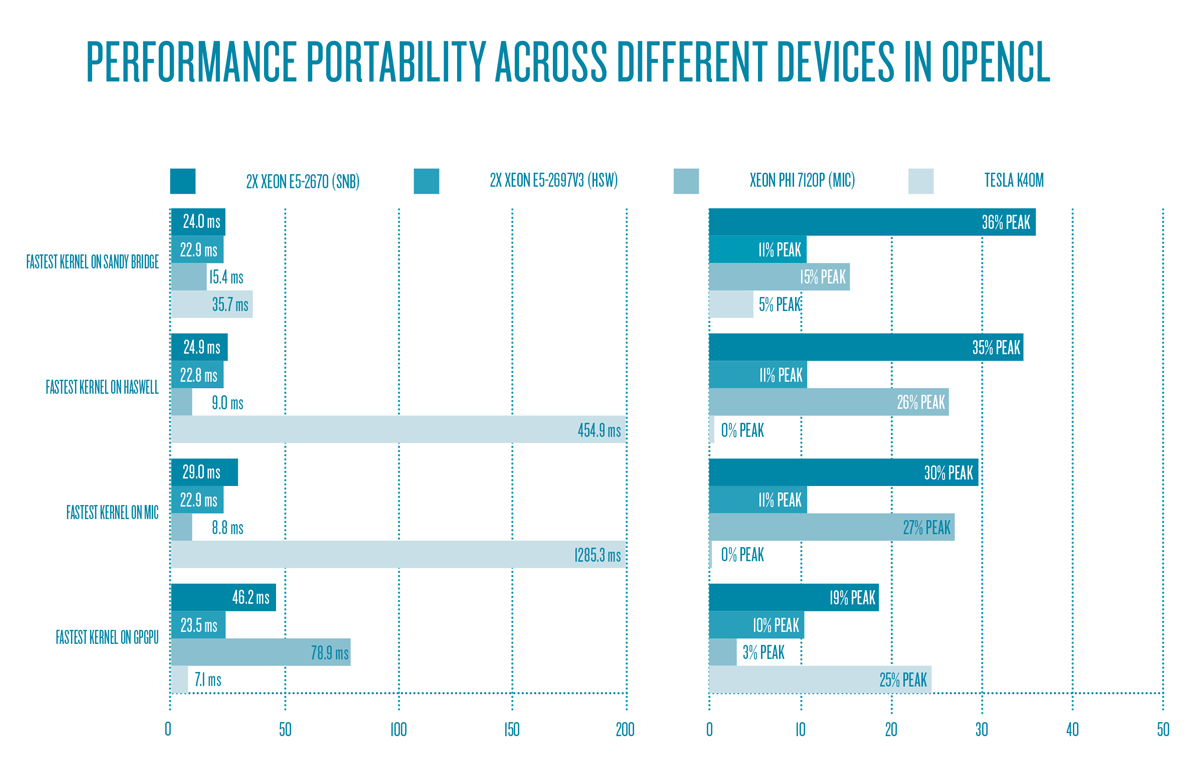
Figure 6: Runtime and device utilization of the best-performing kernels of each device across devices – performance is not portable.
VISUAL ANALYSIS OF ENERGY FLOW
The majority of methods for visual analysis of biomolecular systems is focusing on structural data, resulting from classical molecular dynamics simulations, for example. These methods need to be extended to reveal the extent of quantum-
mechanical coherences shared between different chlorophylls. For this task, a ZIB-internal collaboration (bridge project) between the Distributed Algorithms and Supercomputing department and the Visual Data Analysis department was formed.
The goal is to develop novel visual methods and tools for analyzing:
a) Time-dependent quantities and flows through a molecular network; and
b)The spatiotemporal extent of quantum-mechanical coherence.
This includes:
- A combined visual representation of spatial molecular structure and a related directed graph with time-dependent attributes;
- The identification of dominant pathways and causes for delays by extracting flux information from time-dependent populations of network nodes; and
- Time-dependent tracers for revealing quantum-mechanical superposition states.
The resulting techniques will be integrated into the publicly available exciton dynamics tool previously developed by Kramer/Kreisbeck at nanoHUB.org.
The insights gained from the analysis of the energy flows are also expected to impact the simulation itself by revealing a pattern that allows to filter nodes inside the hierarchy graph that have a negligible impact on the result. This would help to further reduce the needed computational resources and allow for an improved time-to-solution or deeper hierarchies.
Collaborations
Prof. Alán Aspuru-Guzik, Harvard University, Department of Chemistry and Chemical Biology works on the quantum chemistry of photosynthetic complexes.
Prof. Thomas Renger, Johannes-Kepler Universität Linz, Institut für Physik, provides parameter sets for various photosystems.
Prof. Gerhard Klimeck is the Director of the Network for Computational Nanotechnology which powers nanohub.org at Purdue University.