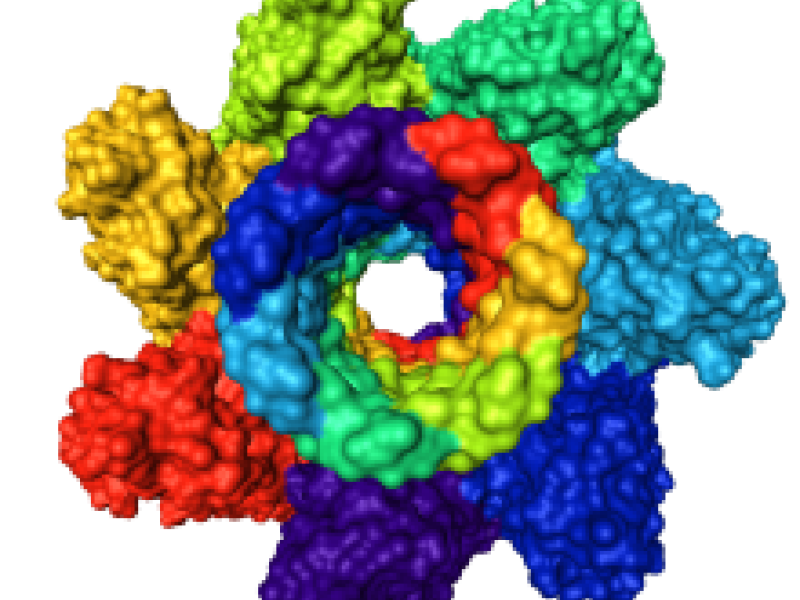
The understanding of biochemical processes requires the study of structure and dynamics of biomolecules of varying size. In these investigations vast amounts of data are generated both in experiments and in numerical simulations. An interplay between chemical classification, mathematical data reduction and graphical data representation is necessary to aid in uncovering the essential structure of biomolecular processes and making them understandable to the human observer. The aim of this project is to develop a software environment which integrates molecular dynamics simulation, visualization, and analysis.