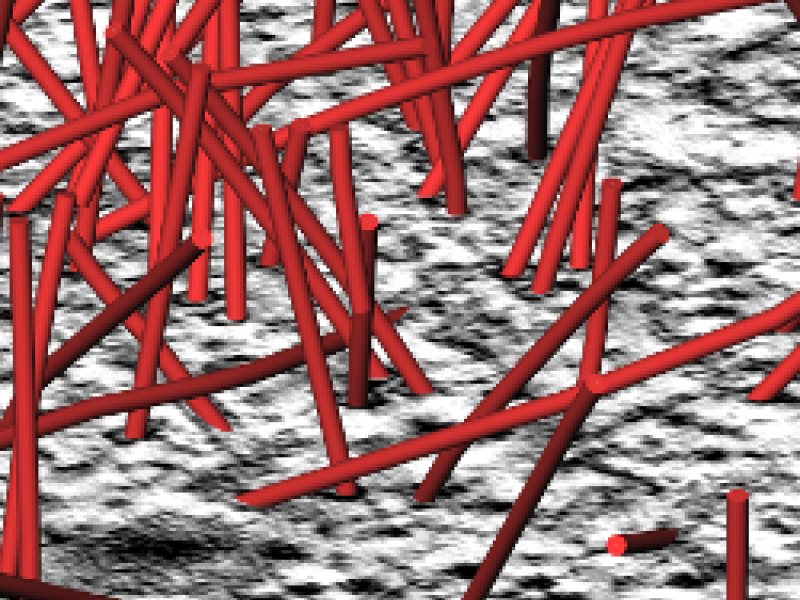
Microtubules are a part of the cytoskeleton of cells. They are tube-like polymeres that stabilize the cell but also play an important role in cell division. In particular the spindle - the microtubule assembly formed during cell division - is subject to intense research. One of the most important methods for looking at microtubule distribution in cells is using electron tomography. Here thick (300nm) sections of cells are prepared for electron microscopy and imaged on a CCD camera. Inverse radon transform algorithms are then used to reconstruct 3D information. One of the main bottle necks in the analysis of the microtubule network is then the construction of an exact model of the microtubules from the acquired data. This requires laborious manual modeling of microtubules. The aim of this project is to utilize image processing techniques to significantly accelerate the reconstruction of microtubules in the sections and thus reduce manual operator time and collect data in a way that is less biased by human operations.