This page describes how to obtain the Filament Editor software and related modules for reconstruction, and morphometric analysis of (particularly in vivo labeled) neurons as described in [1]. The software was developed at Zuse Institute Berlin (ZIB), the Max Planck Florida Institute (MPFI), and the Max Planck Institute for Biological Cybernetics (MPI-BC). It is based on and requires Amira (FEI Visualization Sciences Group, FEI-VSG). Contributions of FEI-VSG to the Filament Editor are marked as such in [1].
To use the software, do the following:
- Obtain Amira 5.5 or Amira 6.0.1. For users without Amira, a trial version of Amira together with a time-limited license can be applied for from FEI-VSG.
- Download and install the ZIB extension package from here. The extension package is available for Windows 64-bit, Linux 64-bit, and MacX (Amira 6.0.1 version only). The extension package is subject to the license contained in the package (LICENSE.txt).
Amira 5.5 and its extension package have been used to create the results in the paper [1]. The extension package for Amira 6.0.1 contains minor changes.
A brief howto on how to install the software and use it to reproduce results published in [1] can be downloaded from here.
Also, we have made available the data used in the paper. It can be downloaded from here (~410 MB). If you use any of the data in your own publication, please cite appropriately (see README.txt in the data package for details).
Some key features of the Filament Editor are illustrated in the following movies:
- Splicing [MPG, 39 MB]. The Filament Editor is used to proof-edit a fragmented branch. Using camera rotation in the 3D viewer the user determines the fragments to splice, and accomplishes this by selecting the branches and invoking the splice operation. Tracing artifacts are cut off by connecting a node to an edge point and deleting the resulting spur. All intermediate nodes are removed in a final step, resulting in a single connected branch.
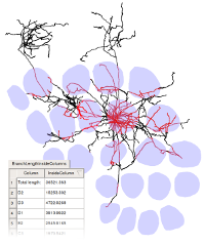
- Semantic labeling and morphometric analysis with respect to reference structures [MPG, 45 MB]. A Layer 5 slender-tufted neuron in rat vibrissal cortex has been completely reconstructed and semantically labeled using the Filament Editor. Substructures can be quickly selected using the Label View. Temporarily hiding selections (landmarks in this case) allows to focus on remaining regions (neuron morphology). To quantify the total axon length within the D2 column, the user selects a D2 barrel contour and the axon. The CorticalColumn analysis tool virtually extrudes the contour and computes the contained length of the selection, presented to the user in a spreadsheet. The contained points are colored for visualization. A similar analysis is available for surfaces generated from contours.
- Alignment [MPG, 32 MB]. Eight tracings from adjacent sections are merged into a single SpatialGraph and aligned using the automatic alignment method integrated in the Filament Editor. Coloring matched pairs of nodes facilitates the splicing across section boundaries, illustrated for one branch.
Reference
[1] Vincent J. Dercksen, Hans-Christian Hege, Marcel Oberlaender (2014): The Filament Editor: An Interactive Software Environment for Visualization, Proof-Editing and Analysis of 3D Neuron Morphology. NeuroInformatics 12(2), pp.325-339. DOI