CONTROLLING RISK EMERGING FROM DRUGS IN THE WATER CYCLE
After passing through our body remnants of drugs get into the water cycle. The largest part of these drug wastes are degraded in water purification processes. However, traces of the drugs themselves and in particular their transformation products can still be found in our drinking water and constitute a significant threat to our health. Systematical research on the transformation process has started only a few years ago. Just recently first ideas of how to save the water cycle from potential risks have been presented.

ANTIBIOTICS RESISTANCE
Since 2011, Vedat Durmaz is working at ZIB for the research project “TransRisk” funded by the Federal Ministry of Education and Research. TransRisk deals with the characterization, communication and minimization of risk emerging from new pollutants and transformation products in the water cycle. Together with Harald Mückter at LMU Munich, about 30 transformation products have been identified for the antibiotics sulfamethoxazole SMZ (Fig. 2). Transformation products may also have an antibiotic effect and, thus, can cause the increase of antibiotic resistant bacteria
“With our computational strategy it is possible to prioritize the transformation products for further toxicological investigations”, Vedat Durmaz says. The aim is, e.g., to compare the 30 transformation products with SMZ and to estimate which one has similar effects on the essential bacterial target protein. Marthe Solleder, a student of Vedat Durmaz, translates the molecular structures of the transformation products into physical models in the computer. On the basis virtual screening of all possible transformation products via molecular simulations are possible (FN:V. Durmaz, S. Schmidt, P. Sabri, C. Piechotta, M. Weber {2013}. A hands-off linear interaction energy approach to binding mode and affinity estimation of estrogens. J. Chem. Inf. Model, 53{10}:2681–2688.), (FN:V. Durmaz, M. Weber, R. Becker {2012}. How to Simulate Affinities for Host-Guest Systems Lacking Binding Mode Information: Application in the Liquid Chromatographic Separation of Hexabromocyclododecane Stereoisomers. Journal of Molecular Modeling, 18{6}:2399-2408.). By this strategy, previously unknown transformation products have already been identified that exhibit significantly strong antibiotic effects but are hardly degradable by water cleaning technology.
ESTROGENE EFFECTS
The virtual screening concept has already been successful for the estimation of estrogenic effects caused by transformation products. In this case, the target protein is the human estrogene receptor. In a cooperation with Christian Piechotta from the Federal Institute for Materials Research, the relative binding affinities measured in the laboratory were compared with the results from molecular simulations. “We found a very high correlation between these two quantities”, Marcus Weber, leader of the ZIB working group, says (FN:V. Durmaz, S. Schmidt, P. Sabri, C. Piechotta, M. Weber {2013}. A hands-off linear interaction energy approach to binding mode and affinity estimation of estrogens. J. Chem. Inf. Model, 53{10}:2681–2688.).
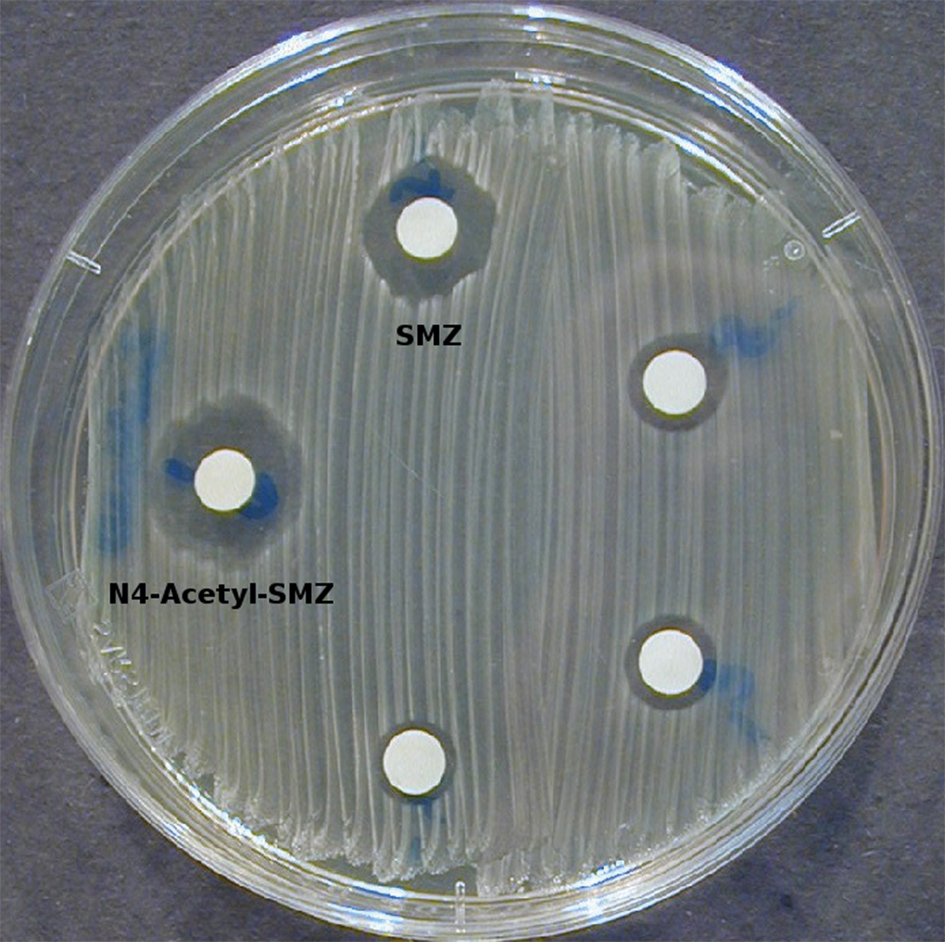
A simple test in laboratory shows that a transformation product of SMZ has an antibiotic effect. This transformation product can be found in the water cycle and is less degradable compared to SMZ.
TWEAKING MOLECULAR SIMULATIONS FOR MANY-CORE PROCESSORS
As described above, the simulation of drug-like molecules and the prediction of their interaction properties are of great interest. Unfortunately, for these small-sized molecular species, traditional computational approaches fail to benefit from many-core processors simply due to their insufficient problem size. Even if one considers molecules in its aqueous environment, when solvation effects have to be taken into account, all-atom simulations including sufficiently many water molecules tend to be still too small-sized to exploit the computational capabilities.
CONCURRENT KERNEL EXECUTION
One possible solution for modern many-core processors is to utilize the feature of Concurrent Kernel Execution, meaning that multiple computational tasks are offloaded to the many-core device so that a higher resource utilization and significant speed-ups can be achieved. Thomas Steinke, the head of the supercomputing group at ZIB, explains a problem with this approach: “Although the Concurrent Kernel Execution feature is offered on the Nvidia Fermi GPU architecture we identified weaknesses in its implementation.” Together with cooperation partners his group developed an alternative approach to allow for a higher computational throughput for molecular simulations (FN:F. Wende, F. Cordes, T. Steinke {2012}.On Improving the Performance of Multi-threaded CUDA Applications with Concurrent Kernel Execution by Kernel Reordering, in Proceedings of the 2012 Symposium on Application Accelerators in High Performance Computing, SAAHPC ’12, {Washington, DC, USA}, pp. 74–83, IEEE Computer Society, 2012.). The software GLAT (FN:F. Cordes {2013}. GLAT – Global Local Adaptive Thermodynamics.” GETLIG&TAR.) is based on a new idea of how to compute the interactions of surrounding water molecules on many-core processors (see Fig. below).
HIGH SPEED-UP FACTORS
“We continued our architectural analysis on today’s many-core platforms NVidia Kepler GPU and Intel Xeon Phi”, Thomas Steinke says. His group demonstrated improvements of vendor architectures and the advantages of their software approach (FN:F. Wende, F. Cordes, T. Steinke {2012}.On Improving the Performance of Multi-threaded CUDA Applications with Concurrent Kernel Execution by Kernel Reordering, in Proceedings of the 2012 Symposium on Application Accelerators in High Performance Computing, SAAHPC ’12, {Washington, DC, USA}, pp. 74–83, IEEE Computer Society, 2012.), (FN:F. Cordes {2013}. GLAT – Global Local Adaptive Thermodynamics.” GETLIG&TAR.). “With two Xeon Phi processors we can handle twice the number of simulations with almost no increase in the overall execution time”, he explains the advantages of the new approach which helps researchers in the CMD group to approach more advanced problems.
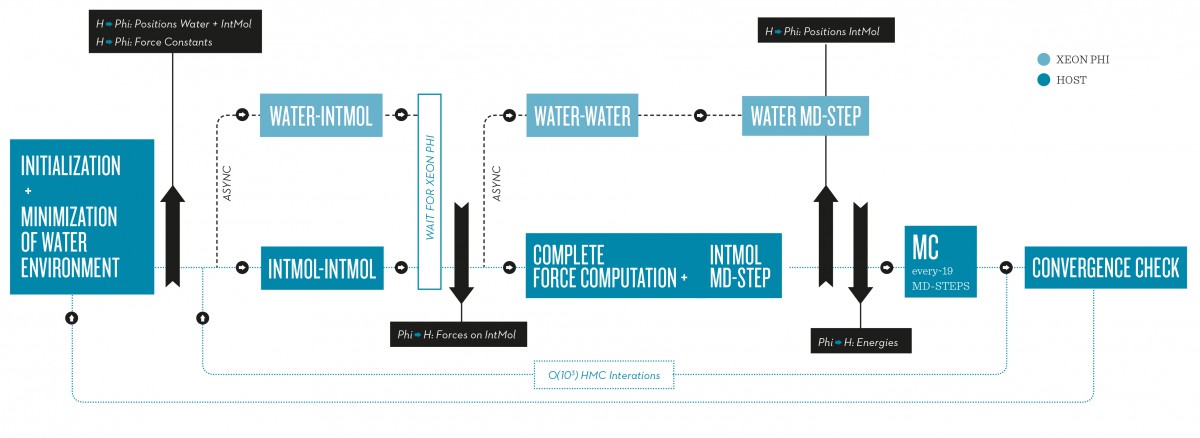
Heterogeneous computer architectures with many-core processors like general-purpose graphics processors (GPU) or Xeon Phi provide data processing capabilities which are an order of magnitude larger than today’s standard multi-core CPUs. This computational potential can be profitably unlocked for molecular simulations if the implemented algorithms are highly parallel and the problem size suitable large. Otherwise, these resources cannot be fully utilized and, thus, speedups are rather unlikely.
COMPUTATIONAL DRUG DESIGN
Small molecules not only lead to undesirable effects. Our new molecular simulation methods are also able to investigate new drug candidates for clinical research. The CMD working group at ZIB has successfully invented a new pain relief drug concept.

Opioid and mu-opioid-receptor within its aqueous-membrane surroundings
DESIGN OF PAIN RELIEF DRUGS
Opioid agonists, like the well-known morphine, are essential analgesics in the treatment of moderate up to severe pain. Prof. Stein from Charité Berlin explains the problem: “Systemically applied conventional opioid agonists activate opioid receptors (Fig. 1), resulting in analgesia. The use of currently available opioids, however, is limited by major adverse side effects such as the central or intestinal effects (FN:E. Freye {2010}: Opioide in der Medizin, Springer, 8. Auflage.), (FN:Zöllner, C. Stein {2007}: Opioids, Handb. Exp. Pharmacol. {177}:31-63.).” The aim of a BMBF cooperation project is to design opioids acting selectively outside the brain and gut to avoid such side effects.
Olga Scharkoi (Fig. 2) from the Computational Molecular Design working group describes the idea: “We want to activate peripheric opioid receptors only in inflamed tissue. This tissue has a lower pH-value than healthy tissue.” By the use of conformation dynamics methods, three opioid candidates have been proposed, which are presently undergoing successful in-vitro and in-vivo experiments at Charité (FN:C. Stein, M. Weber, O. Scharkoi, P. Deuflhard. Method and system for identifying compounds that bind and preferably activate a target opioid receptor in a pH-dependent manner. EP261327), (FN:C. Stein, C. Zöllner, M. Weber, O. Scharkoi. Fentanyl derivatives as pH-dependent opioid receptor agonists. EP2559685).

Publication in »Bild der Wissenschaft« concerning the opiod design.
MECHANISMS OF OPIOID RECEPTORS
Besides the design of new drugs, the low pH-value in inflamed tissue is supposed to have further effects on the opioid receptor and the activation process. Martina Klimm is a fellow of the SALSA graduate school. She points out: “Because the opioid receptor system is rather complex, classical molecular simulation methods are not leading to sufficient results. One has to use our new methods to obtain statistically appropriate amounts of data from molecular simulation.” A lot is known about the rhodopsin receptor which is a very similar G-protein-coupled receptor. The available simulation data for rhodopsin can be reconsidered for the opioid receptor. Martina Klimm describes her work: “The challenge is to derive the global behavior of the opioid receptor from the available local samplings of the rhodopsin receptor. For this we invented direct reweighting strategies (FN:M. Klimm, A. Bujotzek, M. Weber {2011}. Direct Reweighting Strategies in Conformation Dynamics. MATCH Communications in Mathematical and in Computer Chemistry, 65{2}:333-346.).” In order to analyze the huge statistical data in the end, coarse graining methods are needed (FN:K. Fackeldey, M. Klimm, M. Weber {2012}. A Coarse Graining Method for the Dimension Reduction of the State Space of Biomolecules. Journal of Mathematical Chemistry, 50{9}:2623- 2635.). Together with spectroscopic experiments performed at Technical University Berlin, within the group of Prof. Peter Hildebrandt, the results will help to understand the effects of analgesics at different pH-values.
CAVITY DETECTION IN MOLECULAR STRUCTURES
Opioid receptors are located in the cellular membrane. Membrane proteins are often used for the transport of signals or for pumping ions by forming cavities.
The visual reconstructions of the geometrical shape of cavities are of high interest. During the last decades many algorithms have been presented to achieve this. However, the geometrical accuracy of most of these algorithms is often insufficient. Additionally, many techniques are restricted to compute only a small subset of the internal cavities.
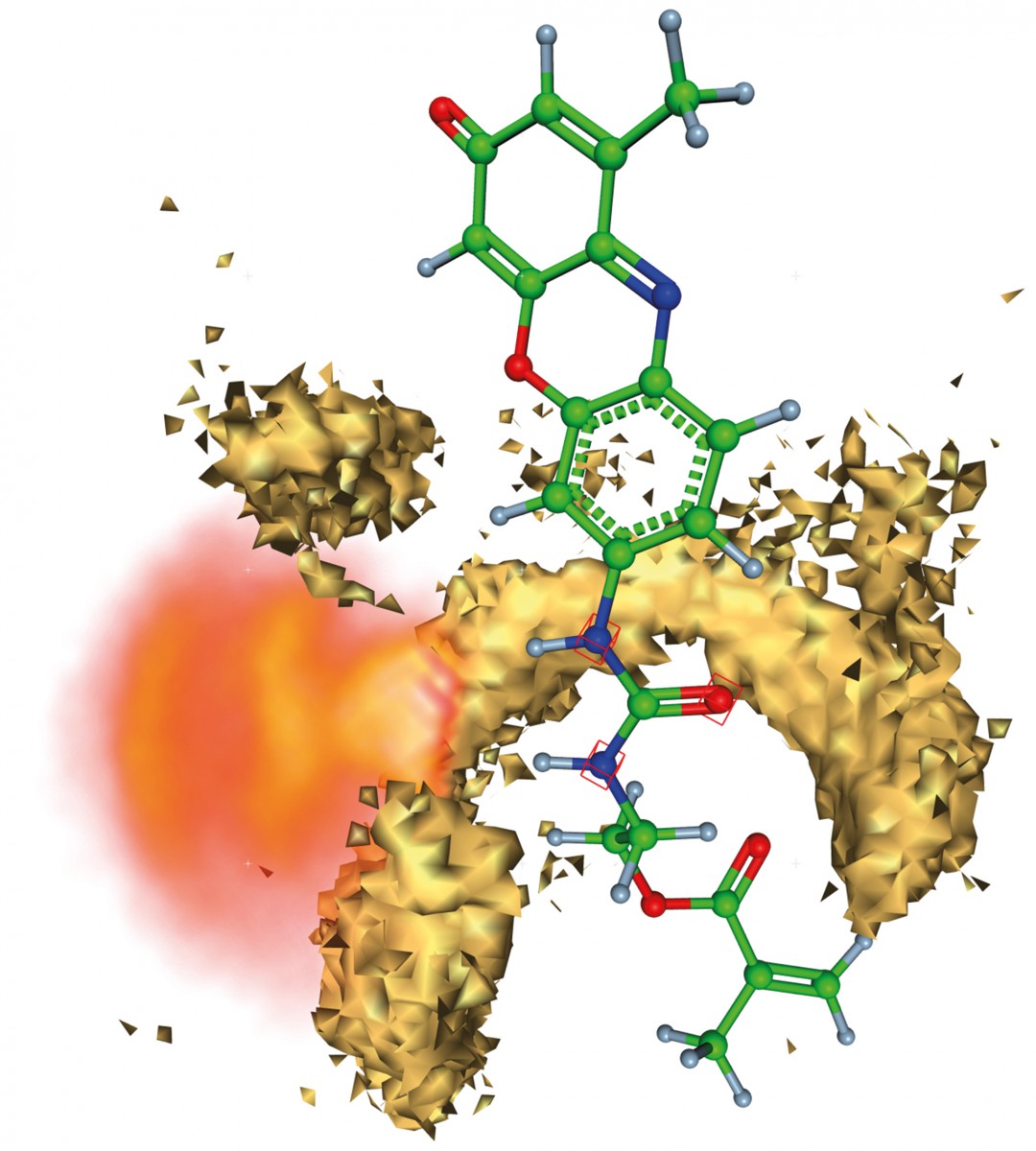
The molecular structure visible in ball-and –stick representation is the template- detecting part inside a MIP. Isosurfaces show the most probable position of polymer atoms which form a specific binding cavity for the template molecule. The flexibility of the template molecules is indicated with a »cloud«.
GENERALIZED VORONOI DIAGRAMS
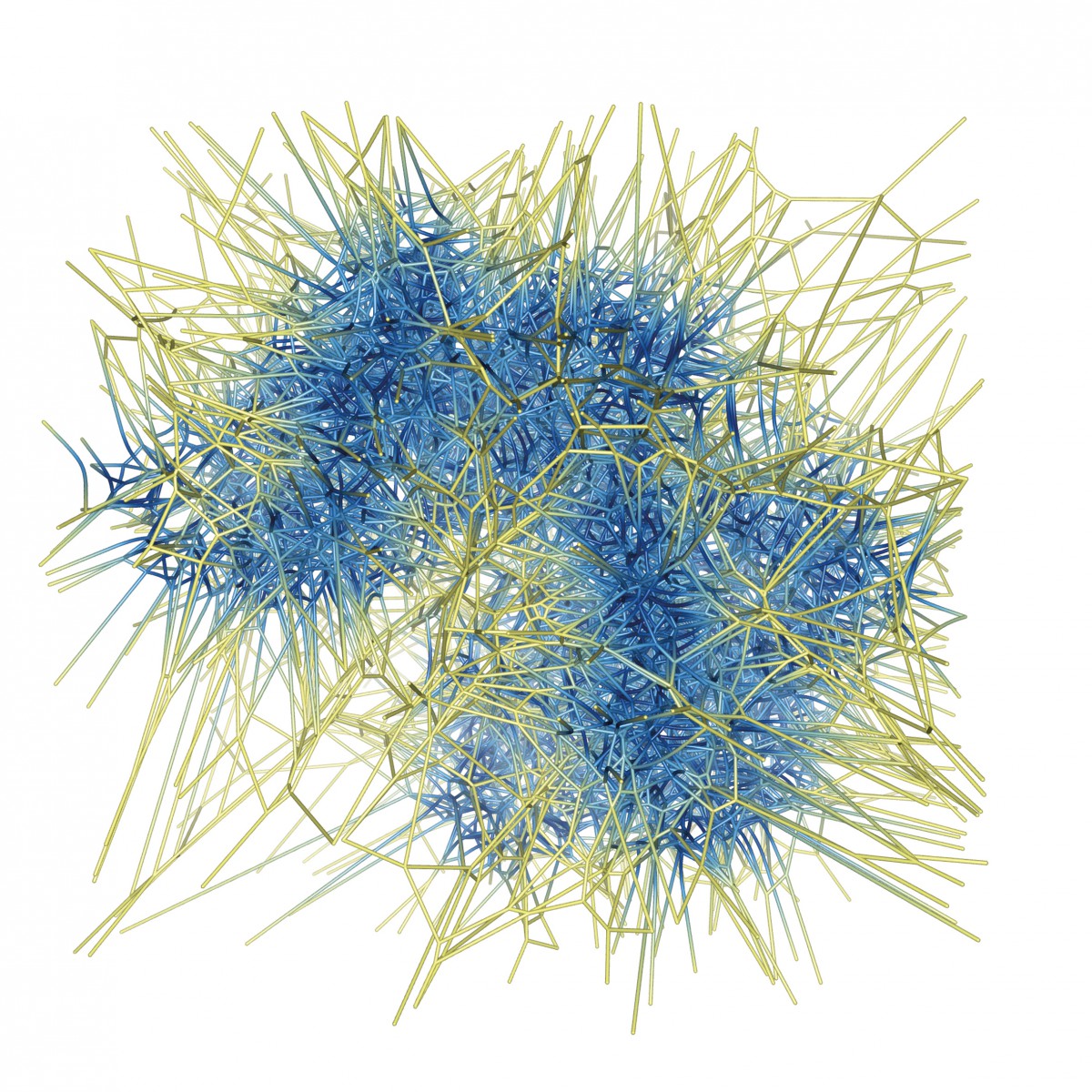
To overcome these limitations, the visualization group of Hans Christian Hege developed a new approach based on the generalized Voronoi Diagram for spheres that represent the atoms (FN:N. Lindow, D. Baum, S. Prohaska, H.-C. Hege {2010}. Accelerated Visualization of Dynamic Molecular Surfaces. Comput. Graph. Forum, Vol. 29, pp. 943-952.), (FN:N. Lindow, D. Baum, H.-C. Hege {2011}. Voronoi-based Molecular Path Detection and Visualization. IEEE Trans. Vis. Comput. Graph., Vol.17, pp. 2025-2034.). Norbert Lindow is a coworker in this group: “We have implemented an efficient algorithm to compute the Voronoi diagram and to extract and visualize all cavities in a molecule at once.”
MOLECULAR IMPRINTING – AN EMERGING TECHNOLOGY
The mechanisms which are responsible for the formation of binding cavities are very important for the studies of molecularly imprinted polymers (MIPs) (FN:B. Sellergren {2001}. Molecularly imprinted polymers. Man made mimics of antibodies and their applications in analytical chemistry. Elsevier, Amsterdam.). “This understanding will facilitate the rational development and the actual preparation of MIPs, in particular, of MIPs with intrinsic signal generation”, says Knut Rurack the cooperation partner from the Federal Insitute of Materials Reseach and Testing: “Since several decades, the concept of molecular imprinting is successfully explored as a synthetic alternative to immunochemical recognition methods (FN:Wagner, W. Wan, M. Biyikal, E. Benito- Peña, M.C. Moreno-Bondi, I. Lazraq, K. Rurack, B. Sellergren {2013}. J. Org. Chem. 78:1377-1389,).” The molecule to be detected by the MIPs is denoted as template molecule. Vedat Durmaz from the CMD working group creates physical models for each proposed polymer mixture: “Each polymer mixture is simulated on the computer twice, once with template molecule in it and once without.” From this computational data the cavity forming quality is derived in order to find the best MIP with the most specific binding of the template molecule.
MOLECULAR CAVITY DYNAMICS
The internal cavities of proteins are dynamic structures and their dynamics may be associated with conformational changes that are required for the functioning of the proteins. In order to study the dynamics of internal protein cavities, appropriate tools are required that allow rapid identification of the cavities as well as assessment of their time-dependent structures. In a cooperaion with Ana-Nicoleta Bondar from FU Berlin (FN:N. Lindow, D. Baum, A.-N. Bondar, H.-C. Hege (2012). Dynamic Channels in Biomolecular Systems: Path Analysis and Visualization. Proceedings of IEEE Symposium on Biological Data Visualization (Biovis ’12), pp 99-106, 2012), (FN:N. Lindow, D. Baum, A.-N. Bondar. H.-C. Hege {2013}. Exploration of Cavity Dynamics in Biomolecular Dystems. BMC Bioinformatics, 14 {Suppl 19}: S5.), the visualization group of Hans Christian-Hege applied their above mentioned static cavity detection algorithm to dynamic data from molecular simulations. “Initially, the cavities are computed for each time step separately”, explains Norbert Lindow: “The new feature allows the user to interactively select a set of cavities in an arbitrary time step and trace them over time.” By this procedure the geometrical overlap of cavities of consecutive time steps is analyzed. Another coworker in the visualization group is Daniel Baum: “Our techniques for visualizing the cavity dynamics in a single image helps to analyze the behavior of the cavity structure. Apart from the shape dynamics one can also analyze topological events like splits and merges by inspecting abstract graph representations.”
ADVANCED MATHEMATICAL METHODS FOR MOLECULAR SIMULATION
Voronoi diagram of possible molecular pathways through a protein structure.
REBINDING EFFECTS
Classically ligand receptor interaction is described using a key-lock analogy: the ligand (the key) seeks for a suitable binding site at the receptor (the lock). “However, this analogy is no longer justified, when considering a real docking process in a thermodynamic environment”, Konstantin Fackeldey from the CMD working group argues: “Here, the ligand and the receptor are flexible and constantly in motion. Moreover, a receptor often has more than one binding site.” The receptors and the ligands can be presented in a multivalent manner, allowing for a simultaneous binding on multiple sites. This kind of binding is investigated together with the Collaborative Research Center CRC 765 located at FU Berlin. In the joint project it has been figured out that the spatial arrangement of multivalent ligands and receptors (FN:M. Weber, K. Fackeldey {2013}. Computing the Minimal Rebinding Effect Included in a Given Kinetics, accepted for publication in SIAM Mult Mod.), (FN:A. Bujotzek, O. Schütt, K. Fackeldey, A. Nielsen, M. Weber {2013}. Efficient Conformational Analysis by Partition-of-Unity Coupling. Math Chem accepted {2013}, http://dx.doi.org/10.1007/ s10910-013-0265-1, also as ZIB-Report 13-58.) lead to faster association and slower dissociation kinetics if the ligands approach the receptors.
One class of efficient simulation methods to model such a binding process are a special class of conformations dynamics algorithms, the so-called Markov State Models, that aim at a partition of the state space into subsets such that the dynamical behavior of the system can be described in terms of transition probabilities which lead to a much better understanding of the processes.
NEW MATHEMATICAL APPROACHES
All conformation dynamics approaches like Markov State Modelling require a Galerkin discretization of the underlying transfer operator. Despite considerable progress in the theory of transfer operators and metastability in the last decade, fundamental problems are still open. Adam Nielsen, working at ZIB as a graduate fellow at the Berlin Mathematical School, the joint graduate school of Berlin’s mathematics institutes, has solved one of these problems: “We were able to show for which classes of ansatz spaces a set-based Galerkin discretization of the generalized transfer operator converges strongly in the L2 norm” (FN:A. Nielsen, K. Fackeldey, M.Weber {2013}. On a generalized transfer operator, ZIB-Report 13-74.).
Besides such rather fundamental results the research at ZIB still aims at novel algorithmic breakthroughs. One of the main algorithmic challenges in MD still is efficient generation of rare event statistics:Most of the available approach to sampling rare event statistics suffers from very large variance and the need to sample very long MD trajectories (FN:Ch. Schütte, A. Nielsen, M. Weber {2013}. Markov State Models and Molecular Alchemy, submitted to Mol. Phys.), (FN:W. Zhang, C. Hartmann, M. Weber, Ch. Schütte {2013}. Importance sampling in path space for diffusion processes, submitted to JCP.). Very recently the CMD group and working groups at FU Berlin have joined forcesto develop a novel approach that allows for the construction of low variance estimators while keeping required trajectories short. The key idea is to use non-equilibrium driving protocols to enforce the rare event under consideration, thus pushing it to shorter time scales. Novel mathematical results show that there is an optimal driving protocol that allows for reproduction of the exact equilibrium rare event statistics via the non-equilibrium sampling with zero variance. Wei Zhang, a postdoc at the CMD group, has even been able to show that suboptimal low variance estimators can be computed using low-dimensional reaction coordinates of the system. “These surprising results are the basis of the development of a new class of rare event simulation algorithms”, Christof Schütte explains the future strategy of ZIB in this field.
