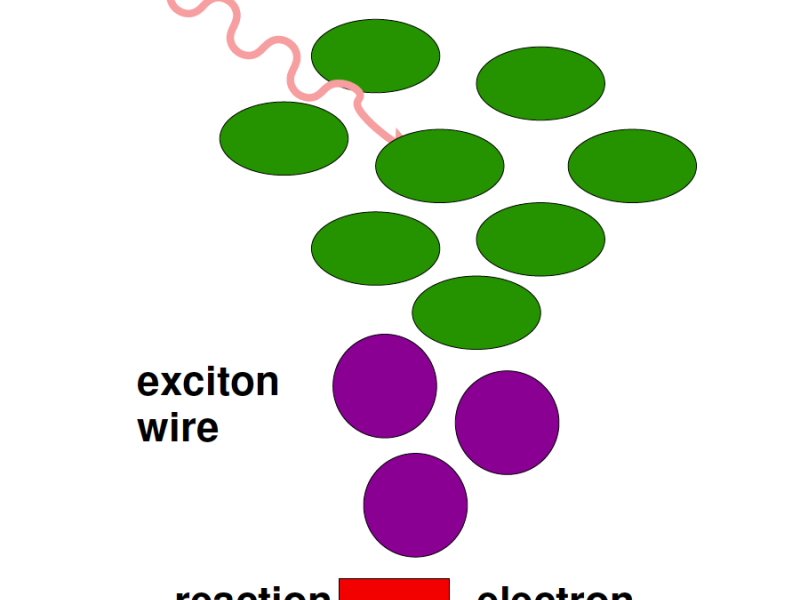
Recent pulsed-laser experiments on molecular complexes yield a time-resolved picture of the energy transport through molecular complexes. The data shows the coupling between electronic and vibrational degrees of freedom with sub-picosecond resolution. Unexpectedly, indications for long-range quantum-mechanical coherences were found and suggest that classical transport models need to be revised to take quantum mechanical effects into account. The realization of “quantum-assisted transport” at ambient room temperature has triggered major research programs to understand molecular networks better and to apply lessons learnt to optimize light emitting OLEDs and organic photovoltaics. To support the analysis of such processes, this project aims at implementing and developing effective visual analysis methods. The majority of methods for visual analysis of biomolecular systems focuses on structural data, resulting e.g. from classical MD simulations. These methods have to be extended to reveal the dynamics of processes and to consider quantum degrees of freedom.
Aims
- Develop novel visual methods and tools for analyzing
- time-dependent quantities and flows through a molecular network
- the spatiotemporal extent of quantum-mechanical coherence
- Integrate novel visualization techniques into the nanoHUB.org exciton dynamics tool developed by Kramer/Kreisbeck