The project aims at a better understanding of the mechanisms responsible for the formation of binding cavities in molecularly imprinted polymers (MIPs). Essential in this regard is a better understanding of the kinetics and thermodynamics of MIPs formation. The project will address the latter by modeling the MIPs formation process in a de novo approach and by verifying the theoretical results by actual synthesis and analytical characterization. Further refinement and improvement of theoretical and experimental steps in iterative loops will approximate MIPs with better controlled sensitivity and selectivity for molecular sensors applications.
The aim of the project is to better understand the mechanisms which are responsible for the formation of binding cavities in molecularly imprinted polymers (MIPs). This understanding will facilitate the rational development and the actual preparation of MIPs, in particular, of MIPs with intrinsic signal generation. Since several decades, the concept of molecular imprinting is successfully explored as a synthetic alternative to immunochemical recognition methods.
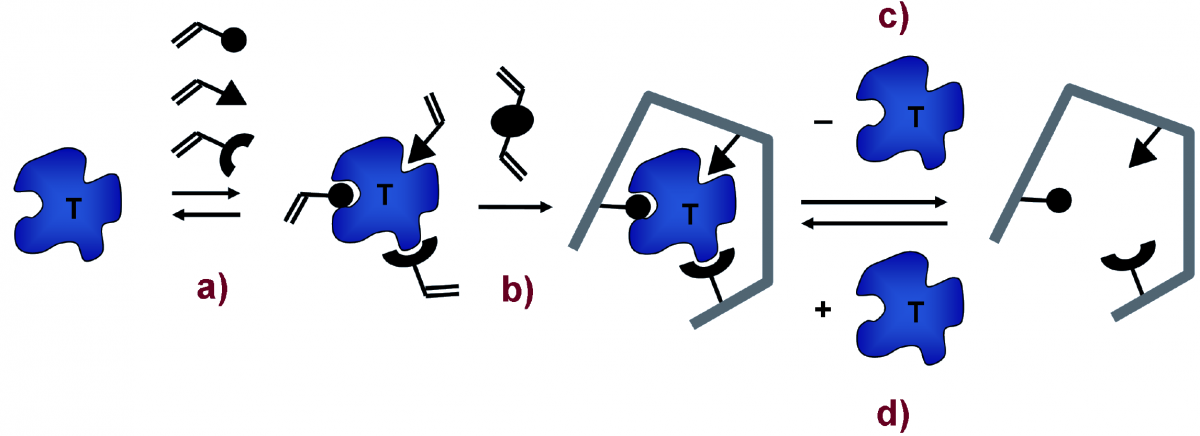
The technique entails the co-polymerisation of functional monomers in the presence of a template, which is removed afterpolymer formation to leave cavities that can be re-occupied by the template in the analytical reaction. The result is a highly cross-linked and durable polymer containing cavities with spe-cific binding sites for the molecule added as template in the polymerisation step.
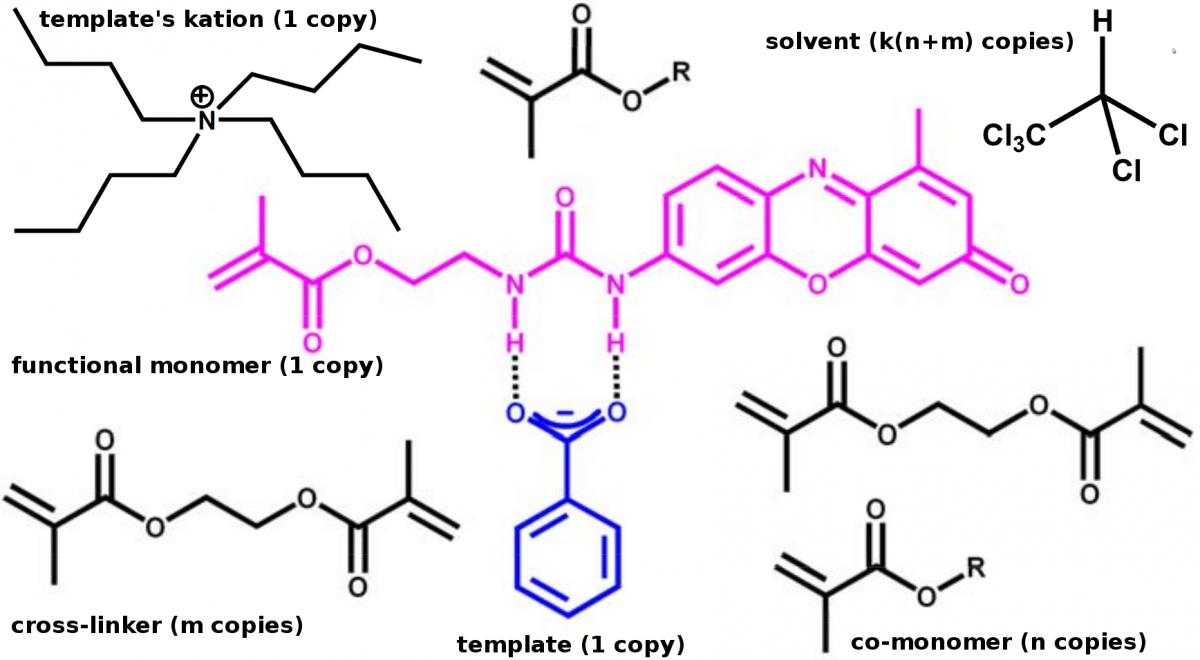
In contrast to the most common approach of empirical synthesis and testing of MIPs, the present project will model the binding scenario by taking into account the template, the functional monomer, possible co-monomers, the cross-linking monomer(s), the solvent or porogen and, if required, certain additives at the quantum level of theory. For this purpose, various chromo- and fluorogenic functional monomers which have been synthesized in the BAM group will serve as starting points for the calculations with particular emphasis lying on their solubility and deprotonation features in polymerisation-relevant solvents. Apparent pKa values as well as specific solvent effects and HBD–HBA interactions will be assessed for model templates by the ZIB group.
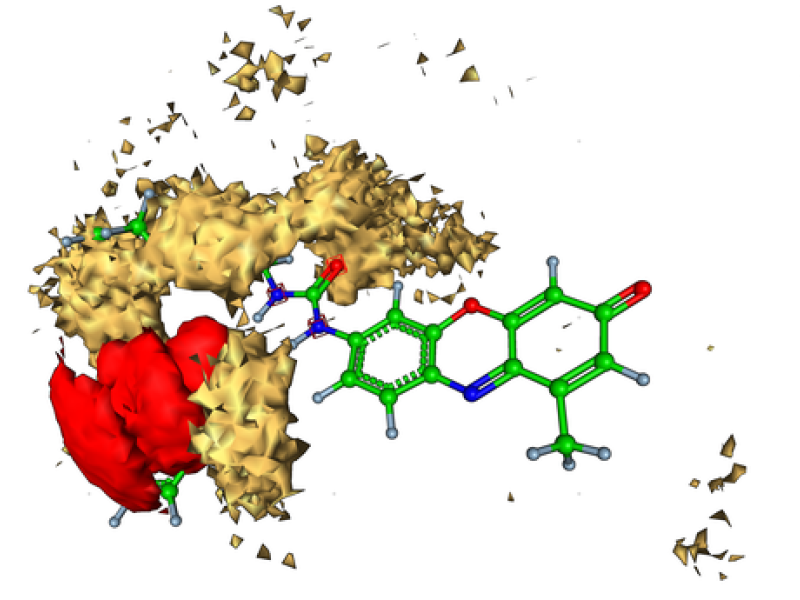
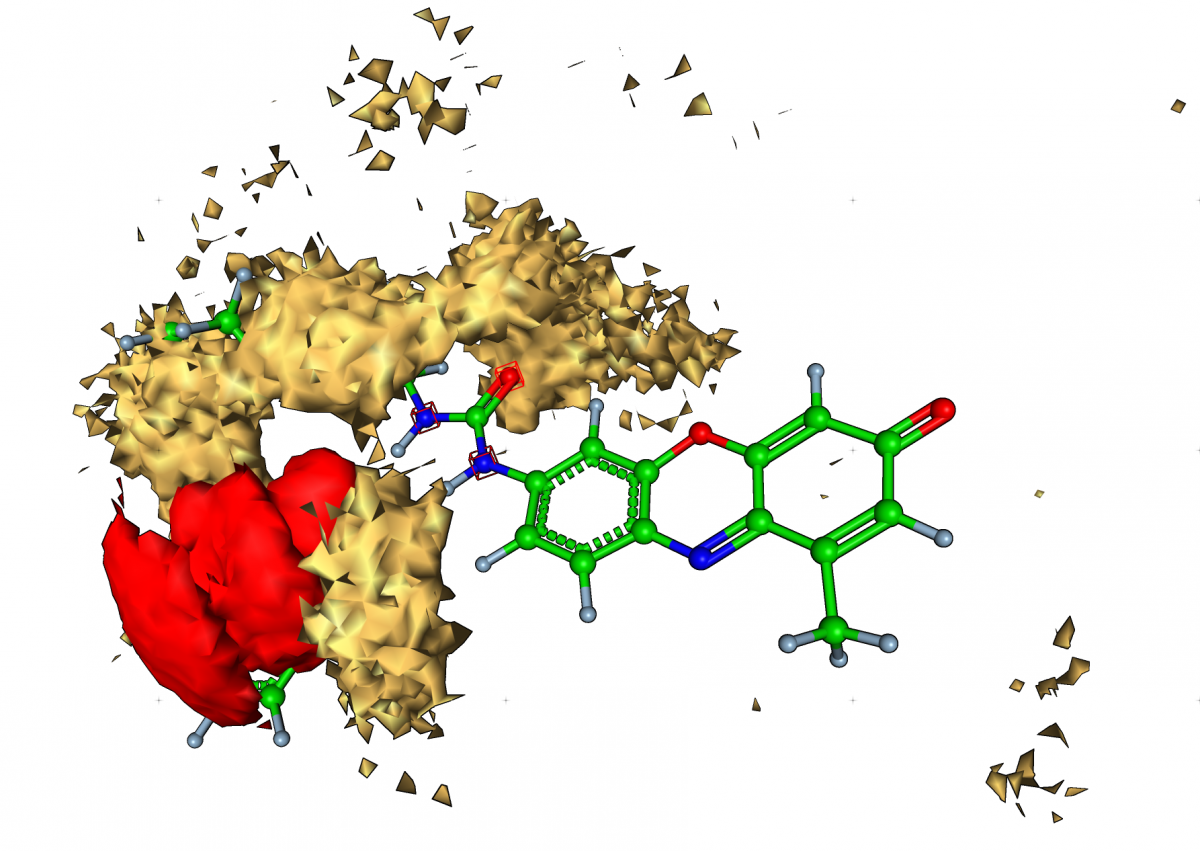 On the right, we have depicted the functional monomer surrounded by some colored objects created on the basis of molecular dynamics data (trajectory with molecular geometries from many time steps). These objects are called isosurfaces and represent those spatial areas where the co-monomers (gold) and the template molecule (red) prefer to reside more often than defined by a cutoff value during the simulation. The aim is to find descriptors from the simulation data correlating strongly with experimental observations which are fluorescence values measured for several mixtures of co-monomers and cross-linkers. In general, each mixture is produced twice, once with the template and once without in order to find unspecific bindings.
On the right, we have depicted the functional monomer surrounded by some colored objects created on the basis of molecular dynamics data (trajectory with molecular geometries from many time steps). These objects are called isosurfaces and represent those spatial areas where the co-monomers (gold) and the template molecule (red) prefer to reside more often than defined by a cutoff value during the simulation. The aim is to find descriptors from the simulation data correlating strongly with experimental observations which are fluorescence values measured for several mixtures of co-monomers and cross-linkers. In general, each mixture is produced twice, once with the template and once without in order to find unspecific bindings.