In recent years, there has been a revolution in the capability to measure and observe the processes of life on many temporal and spatial scales. Simultaneously, our ability to perform extensive simulations of molecular, cellular, and tissue-scale processes has substantially improved and will continue to do so. Research at ZIB is focused on extending this expertise towards spatiotemporal modeling and simulation across the molecular and cellular scales, from pushing time-scale and accuracy barriers, via a combination of the development of novel theoretical and algorithmic techniques for effective molecular or particle dynamics with machine learning, to spatio-temporal hybrid models coupling molecular with extra- and intra-cellular processes. Our projects target seamless integration of new mathematical approaches with incorporation of experimental data, high performance computing, and/or large-scale data analysis, and contribute to finding new strategies in relevant real-world applications like drug design or neurotransmission.
HFSP Chitons
Many organisms utilize a small number of sensory organs and a centralized nervous system (e.g., the paired eyes and complex brains of vertebrates) to acquire and process...
HFSP Chitons
3D morphological analysis of root canals
This research project investigates the improvement of root canal treatment (RCT) by analyzing the relationship between root canal morphologies and treatment failures...
3D morphological analysis of root canals
Virtual Dissection
Advancements in morphology have enabled the creation of virtual fossil replicas that allow non-destructive processing and unprecedented analysis capabilities. However...
Virtual Dissection
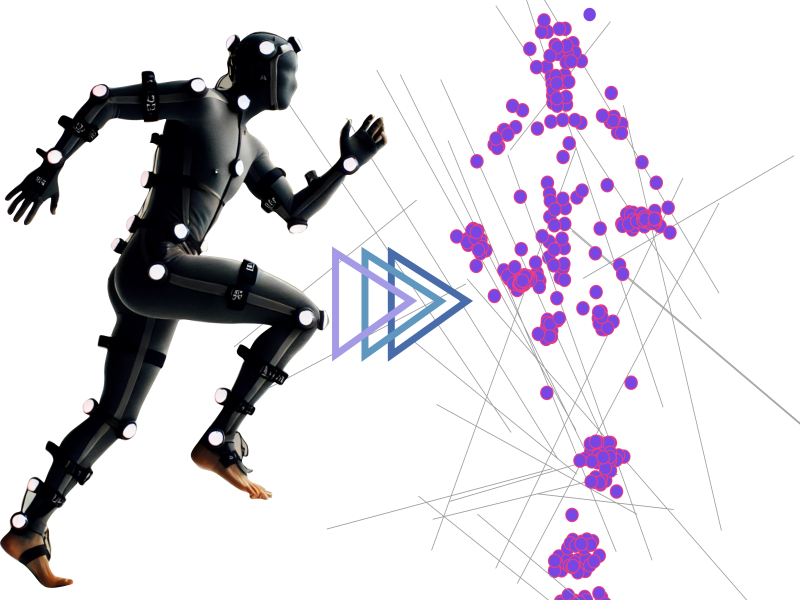
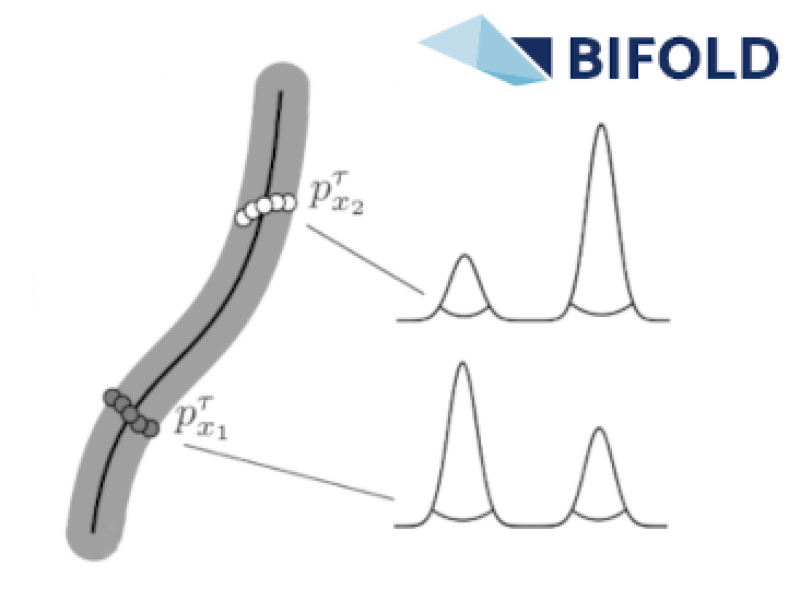
Non-rigid shape registration
Unlocking the next level of 3D shape registration! Our cutting-edge method seamlessly aligns 3D shapes with 2D keypoints captured from multiple cameras, pushing the...
Non-rigid shape registration
HPC and ML for Drug Discovery
High-performance computers are required to simulate complex systems that allow to study the behavior of molecules under real conditions. This project combines artificial...
HPC and ML for Drug Discovery
RobustCircuit
RobustCircuit is a collaborative project of 13 research teams pursuing eight neurobiological projects at five different institutions. The goal of the project is to...
RobustCircuit
Model-Regularized Learning Of Complex Dynamical Behavior
This project is planned to couple machine learning approaches, especially from the field of Deep Learning, with (reduced) ODE models in the sense that the model becomes...
Model-Regularized Learning Of Complex Dynamical Behavior
Geometric Learning for Single-Cell RNA Velocity Modeling
Recent advances in Single-Cell RNA sequencing allow to infer both the gene expression of a cell and the so-called "velocity vector" initializing the changes in that...
Geometric Learning for Single-Cell RNA Velocity Modeling
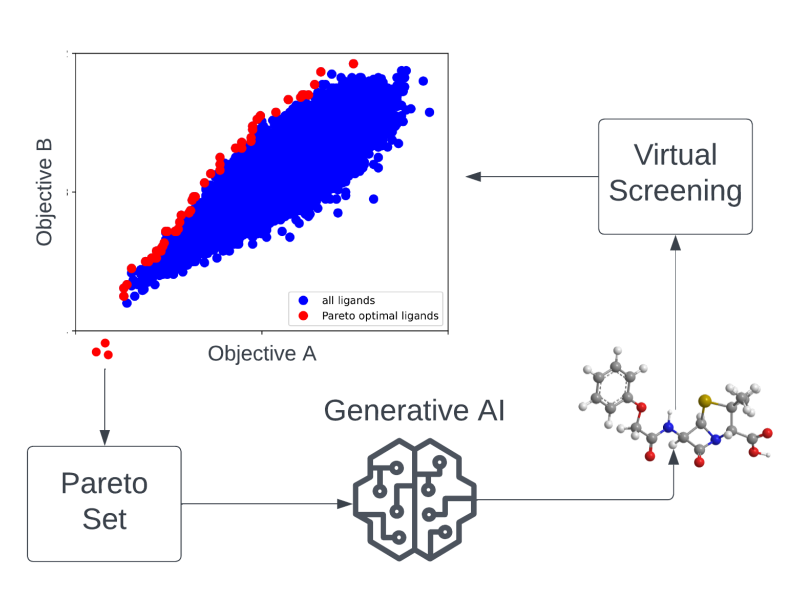
AA1-19 Drug Candidates as Pareto Optima in Chemical Space
The search for novel drug candidates that, at the same time, act with high efficacy, comply with defined chemical properties, and also show low off-target effects can be...
AA1-19 Drug Candidates as Pareto Optima in Chemical Space
Opinion Dynamics
The literature contains a manifold of fundamental models for the dynamical changes of opinions through social influence during social interaction (personal, social...
Opinion Dynamics