This project aims at the creation of a MATH+ success story around the mathematics of drug design. It proposes a new drug design strategy for G-protein coupled receptors by distinguishing between the effects of drugs in healthy and pathological environment. It extends previous projects by (I) coupling the molecular timescales of receptor activation with the associated cellular processes and the adverse side effects induced by them, and by (II) generalizing the new strategy beyond opioid receptors and pain relief towards the treatment of e.g. blood pressure, heart failure, or respiratory failure (e.g. Covid-19).
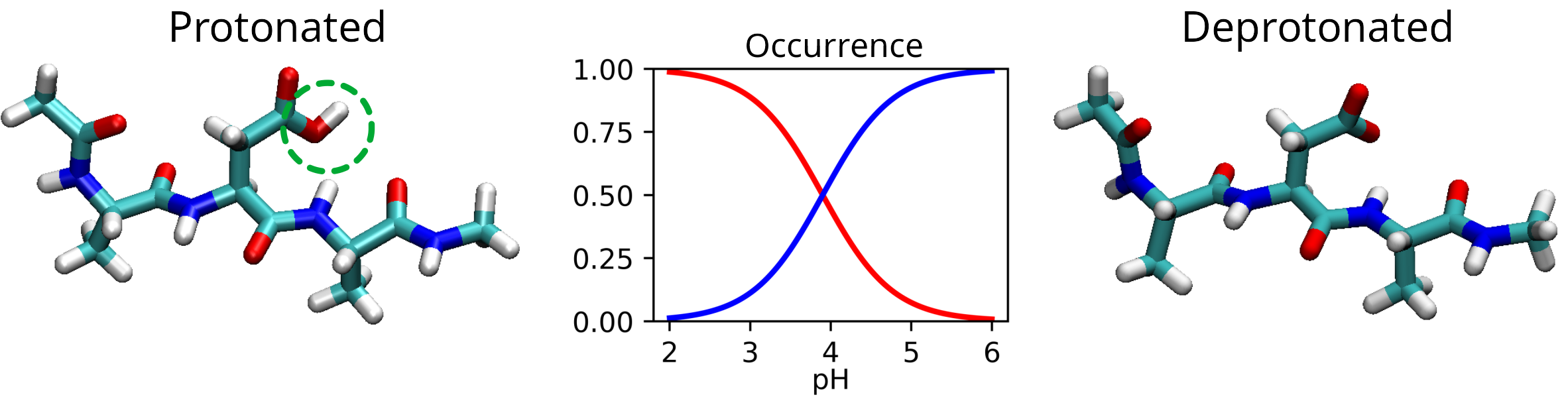
Figure: The tripeptide Ala-Asp-Ala exists in two forms, protonated and deprotonated, whose occurrence depends on the acidity of the environment. Using SqRA, it is possible to determine the transition rate of the peptide as a function of pH.
