The aim of the Computational Anatomy and Physiology group is to improve the understanding of physiological processes, the prediction of disease progress, and the individual planning of complex treatments by mathematical modelling, simulation, and optimization. To this end, we develop efficient adaptive simulation, identification, and optimization algorithms for ODE and PDE problems in several biomedical applications. The resulting algorithms are combined with software prototypes developed in the Computational Diagnosis and Therapy Planning group to form innovative solutions for individualized medicine.
Current research focus are joint diseases, where we compute and identify mechanical loads in joints and optimize implant positions. Further topics are cardiac simulation, forensic medicine, as well as biological and medical image acquisition. Increasing complexity of the biomechanical and physiological processes is gaining importance as we move towards multiscale, multiphysics, and hybrid models, coupling, e.g., multibody mechanics to solid mechanics, or myocardial excitation to mechanical contraction, blood flow and tissue remodelling.
Another important research topic is quantification of uncertainty in models and data, affecting conclusions drawn from simulations as well as parameter identification and optimization algorithms. The algorithms resulting from research in the group are implemented in the finite element toolbox Kaskade 7, a flexible C++ library developed at ZIB. The methods developed in the group also find applications beyond the biomedical field. In collaboration with other research groups at ZIB, we work, e.g., on materials sciences and flight planning.
Example topics
Identification of stresses in heterogeneous contact models
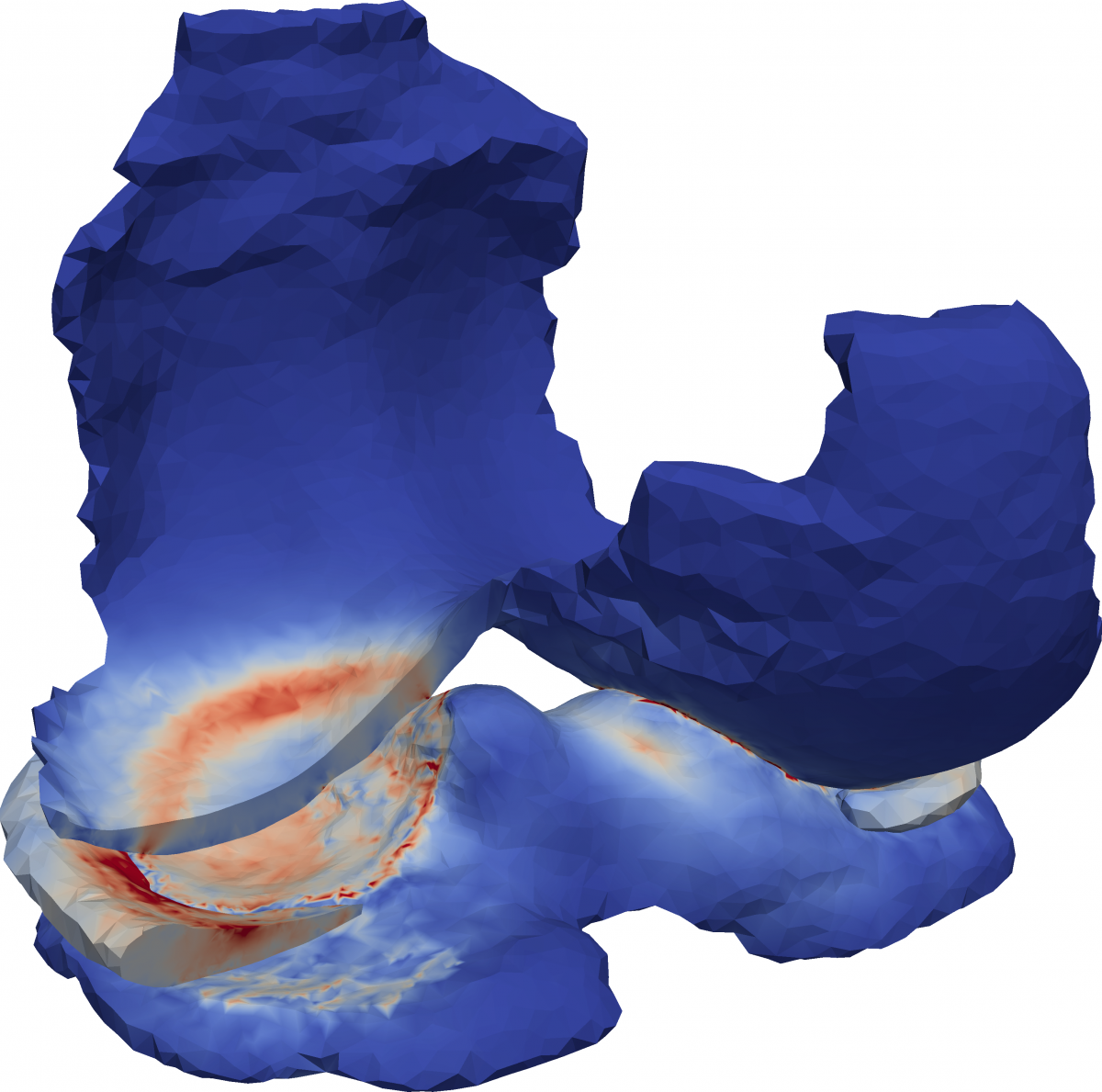 The impact of mechanical loading on osteoarthritis is well known. However, the actual onset and progession mechanisms remain unclear, since local stresses in the joint are inaccessible for measurements. We investigate a coupled model of a biomechanical multibody system and an elastomechanical contact model in the joint for a detailed description of the process. The development of efficient solution algorithms will allow identifying local stresses based on clinical data.
The impact of mechanical loading on osteoarthritis is well known. However, the actual onset and progession mechanisms remain unclear, since local stresses in the joint are inaccessible for measurements. We investigate a coupled model of a biomechanical multibody system and an elastomechanical contact model in the joint for a detailed description of the process. The development of efficient solution algorithms will allow identifying local stresses based on clinical data.
Model-based 4D reconstruction of subcellular structures
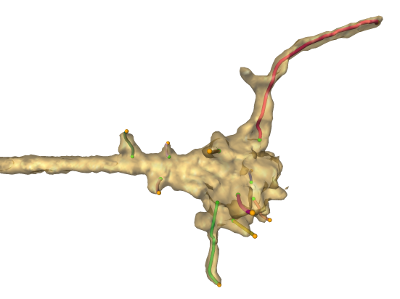 The brain development of flies (Drosophila) is being observed through 4D (3D + time) 2-photon microscopy. The reconstruction of subcellular structures, here filopodia sprouting from a growth cone, is a challenging task currently done semi-automatically with huge manual effort. We aim at reducing the required human labour by using quantitative models of growth cone and filopodia geometry and dynamics for a more robust and consistent algorithmic identification of subcellular structures. Some of the used methods comprise Bayesian inference, optimization, segmentation using convolutional neural networks, and stochastic modelling.
The brain development of flies (Drosophila) is being observed through 4D (3D + time) 2-photon microscopy. The reconstruction of subcellular structures, here filopodia sprouting from a growth cone, is a challenging task currently done semi-automatically with huge manual effort. We aim at reducing the required human labour by using quantitative models of growth cone and filopodia geometry and dynamics for a more robust and consistent algorithmic identification of subcellular structures. Some of the used methods comprise Bayesian inference, optimization, segmentation using convolutional neural networks, and stochastic modelling.
Time of death estimation in forensic medicine
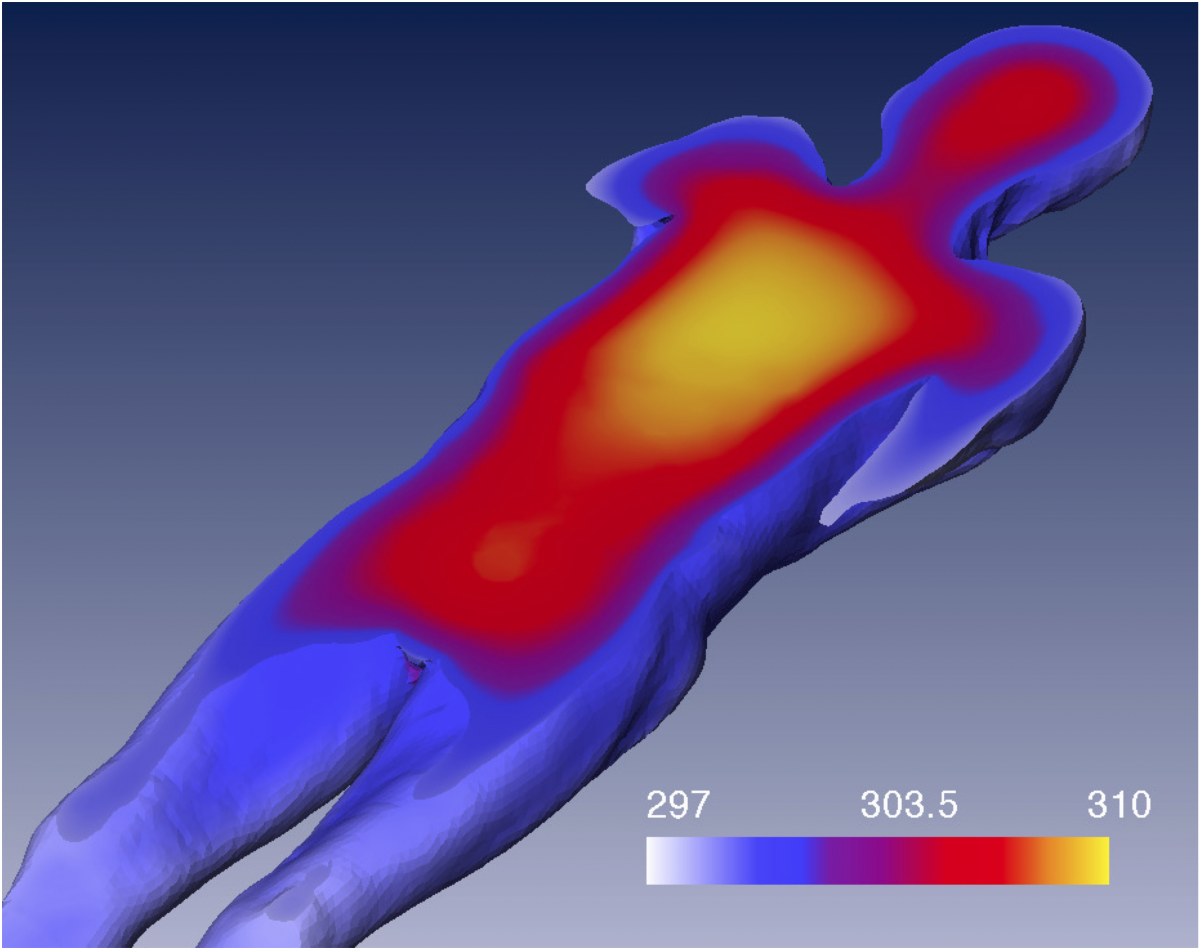 Cooling of corpses is one of the major physical processes from which estimates of the time of death are derived in forensic medicine. We investigate modeling of the physical situation in order to capture the most relevant effects, as well as inversion of the forward problem in order to compute more accurate and reliable estimates.
Cooling of corpses is one of the major physical processes from which estimates of the time of death are derived in forensic medicine. We investigate modeling of the physical situation in order to capture the most relevant effects, as well as inversion of the forward problem in order to compute more accurate and reliable estimates.
Discrete-continuous shortest path problems in flight planning
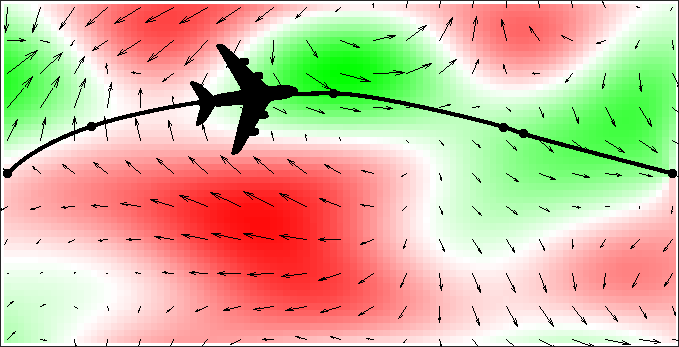 In a growing number of free flight zones, which ideally allow airlines to take any route they prefer, conventional network algorithms are facing a great challenge. As this kind of problem can be solved more efficiently with the tools of continuous optimization, we are investigating how we can incorporate the expretise of this research group and develop novel hybrid algorithms.
In a growing number of free flight zones, which ideally allow airlines to take any route they prefer, conventional network algorithms are facing a great challenge. As this kind of problem can be solved more efficiently with the tools of continuous optimization, we are investigating how we can incorporate the expretise of this research group and develop novel hybrid algorithms.
Group picture
The CAP research group in front of the ZIB building in fall 2021 - and as an example of deep learning style transfer.  G
G
Group picture of department Modeling and Simulation of Complex Processes from summer 2021roup picture of department Modeling and Simulation of Complex Processes from summer 2021
