2D X-ray images play a crucial role for the diagnosis and the therapy planning in orthopaedics. This imaging technique is not only widely available but is also, in contrast to more advanced 3D imaging methods like CT or MRI, considered a fast and inexpensive procedure. However, musculoskeletal anatomy is inherently 3D, and accurate assessment of the patients anatomy is therefore largely dependent upon the individual surgeon’s experience and ability to assess the 3D anatomy by analysis of 2D images only. To overcome this limitation, and in order to enable reliable, quantitative planning of surgical interventions that is less dependent upon the experience of the surgeon, the goal of this project is to provide a method aimed at 3D reconstruction of musculoskeletal anatomy from standard 2D projection X-ray images.
Introduction
Despite the increasing availability of 3D image acquisition methods like computed tomography (CT), conventional X-ray images are still the method of choice for diagnosis and treatment planning in orthopedics. X-rays however only depict a 2D projection of the anatomy of interest, which renders assessing its 3D parameters such as the anatomical shape a challenging task for a human observer (eg. the surgeon). The aim of this project is to develop computer-aided techniques that reconstruct 3D models from a single or a few X-rays, describing both the patient-specific shape and bone density distribution of an anatomy of interest. As an ultimate goal, such patient-specific models will allow for precise joint replacement planning, follow-up and biomechanical analysis, providing essential 3D information solely based on 2D X-rays.
3D-Reconstruction Methods
In previous work (Lamecker et al. 2006, Dworzak et al. 2008, Dworzak et al. 2010, see also THORAX project), we search for a 3D model representation of the anatomy of interest that best depicts the patient-specific 2D shape in the reference X-ray(s). Many variations of the 3D model's shape are projected according to the known X-ray setup, i.e. given a relative position of the X-ray (point) source to the detector. The shape that maximizes the similarity between the model's projection and the X-ray image(s) is assumed to be the best approximation of the true 3D anatomy. Previously, we evaluated the similarity by comparing the projected contours of the model and silhouettes from the reference X-ray. Although contours or silhouettes of 3D anatomical models can be computed very efficiently, they first have to be extracted from the individual patient's X-ray images in a manual or automatic manner. Even more problematic is to use contour information only, as this ignores important information on the interior structures (Fig. 1).
The methods developed in this project aim at overcoming the limitations of previous approaches by regarding as much information contained in the reference X-ray images as possible. Instead of comparing silhouettes or contours, we project virtual X-ray images from the 3D models that simulate a X-ray screening according to the clinical setup. The virtual X-ray images are compared directly to the reference X-rays by means of their intensity distribution, taking the bone-interior structures into account. This way, we increase the robustness of the reconstruction process while determining not only the patient-specific anatomical shape but also its bone-interior density information.
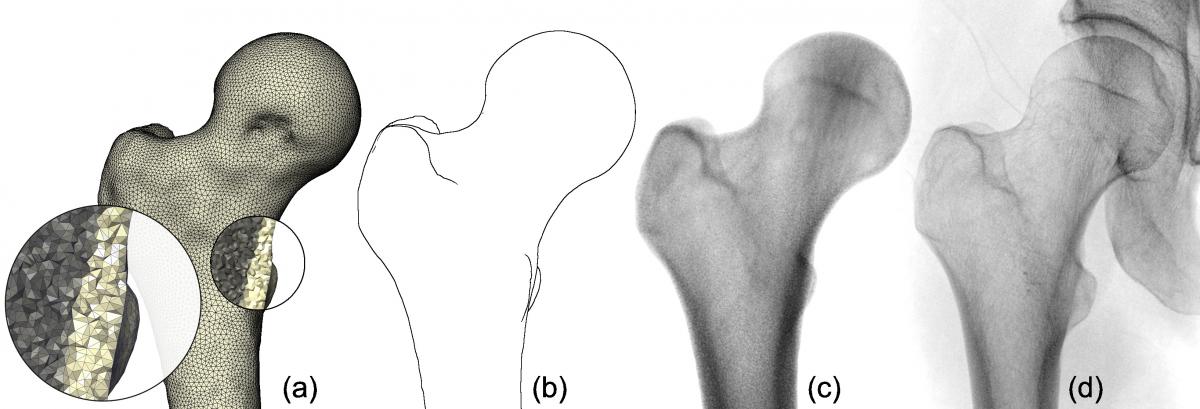
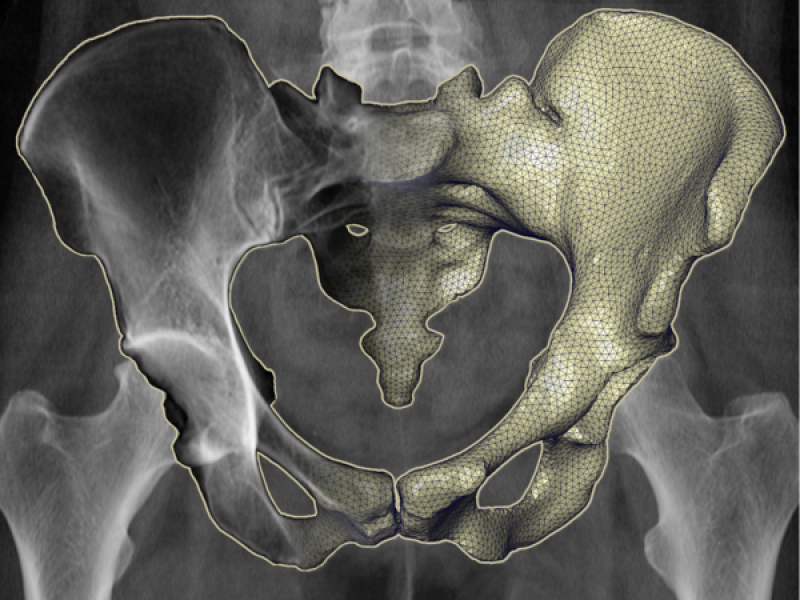
Fig. 1. A 3D model of the proximal femur (upper thigh bone) that incorporates volumetric density information (a). Compared to a projection of contour lines (b), a virtual X-ray (c) depicts the internal density structure of the bone and therefore better resembles the femur in the clinical X-ray (d).
Fast Generation of Virtual X-ray Images
The reconstruction framework has to evaluate many anatomical shape candidates in order to improve the reconstruction quality by identifying the best among several locally optimal solutions. It consequently has to produce large quantities of virtual X-rays (e.g. 10 000 or more images). An essential goal therefore is to quickly compute both variations of anatomical models and their corresponding projections. In addition, the virtual X-rays should depict the density information of the anatomy of interest as accurately as possible to allow for intensity-based comparison with the clinical X-rays.
We developed a hardware-accelerated method that achieves these goals (Ehlke et al. 2013). Compared to previous work, its main contributions are:
- A GPU-algorithm for fast rendering of tetrahedral meshes with higher-order polynomial density functions that represent the shape and density distribution of anatomical structures. Our approach simplifies the tessellation of tetrahedra and can be implemented on the GPU without any explicit branching or looping.
- A method that varies (deforms) both anatomical shape and density information on the GPU and concurrently generates virtual Xray images from the deformed anatomy model.
We evaluated the GPU-algorithm and compared to previous hybrid methods, where the model is deformed on the CPU and projected using the GPU. When regarding more than 40 deformation parameters and higher polynomial degrees of the density function (d = 2 and d = 3), our GPU approach is 6 to 7 times faster than a multi-threaded deformation on the CPU that does not account for the projection time. A comparison of the GPU approach to a hybrid CPU/GPU implementation reveals a performance gain of even 8 to 9 times in the same scenario. We further performed a quality evaluation indicates that our method generates virtual X-rays similar to ground truth images from CT (see Fig. 2).
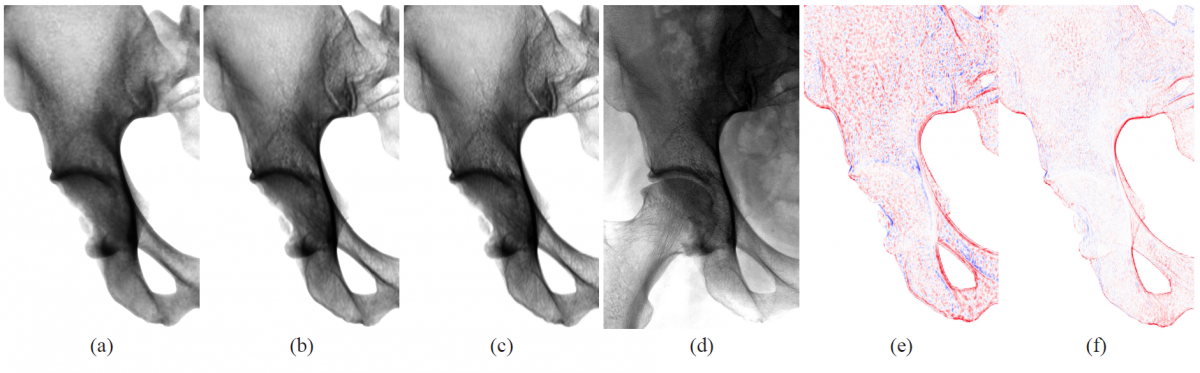
Fig. 2. Comparison of virtual X-ray images to ground truth data. The projections (a) and (b) show close-ups of the right ilium (pelvis) and were generated by applying our method to a tetrahedral mesh of 252k tetrahedra and density function degrees d = 0 and d = 3 respectively. A ground truth projection from CT (c) and a clinical X-ray image (d) of the same pelvis are given for reference. Images (e) and (f) depict the differences of the virtual X-rays (a) and (b) to the ground truth (c) with red indicating positive error, blue negative error.
Anatomical Modelling for 3D-Reconstruction
The reconstruction is performed by searching among a large set of candidates (variations) for the shape that best represents the patient-specific anatomy. It is crucial that anatomically “meaningful” shape candidates are selected, since an anatomically impossible shape might have a similar 2D appearance than the real, patient-specific anatomy and the similarity evaluation in 2D space might thus lead to wrong conclusions. Finding the “right” shape from one or few X-ray images is an ill-posed problem. We therefore introduce a-priori statistical information about the anatomy of interest that acts as a prior during reconstruction, restricting the search space to anatomically meaningful representations.
Previously, statistical shape models (SSMs) proved to be an adequate prior in medical registration problems such as 3D segmentation of CT- and MRI data (Seim et al., 2008, see also Atlas-based 3D Image Segmentation). SSMs utilize the Principal Component Analysis (PCA) to express statistically significant anatomical variations within a training population. We enrich SSMs with volumetric bone-interior information learned from CT and utilize these Statistical Shape and Intensity Models (SSIMs) as a prior for the reconstruction process. The problem of finding the right shape is thus translated into deriving a statistical instance of the SSIM, such that the patient-specific anatomical shape and bone-interior density depicted in the reference images is approximated best.
A treatment plan in orthopaedics does often not only involve single bony anatomies, but rather whole joint structures (e.g. in total hip replacement). The individual components of the joint are oriented in various poses, depending on the patient's physical condition and pose in front of the X-ray scanner. To cope with different joint postures in the reference X-ray images, we further extend the SSIMs to articulated statistical shape and intensity models (ASSIMs). The ASSIM of the hip developed at ZIB represents the hip cup as a ball joint, in which each proximal femur (upper thigh bone) rotates with three degrees of freedom (see Fig. 3). The rotational center of the hip joint is part of the statistical analysis of the model and learned from the training population.
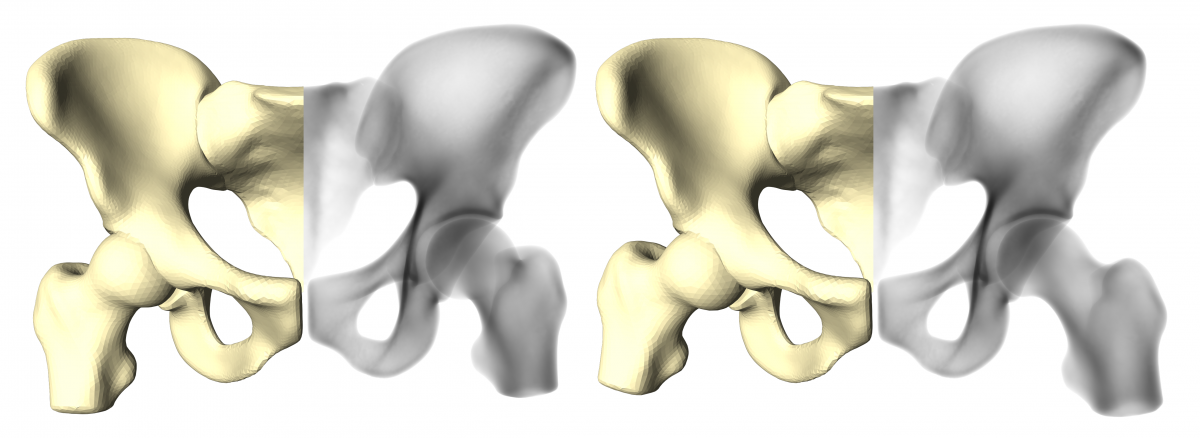
Fig. 3. Articulated Statistical Shape and Intensity Model (ASSIM) of the hip, expressing two different joint postures. Depicted here is the articulated surface of the model (beige) and the bone-interior density distribution (black and white).
Resources