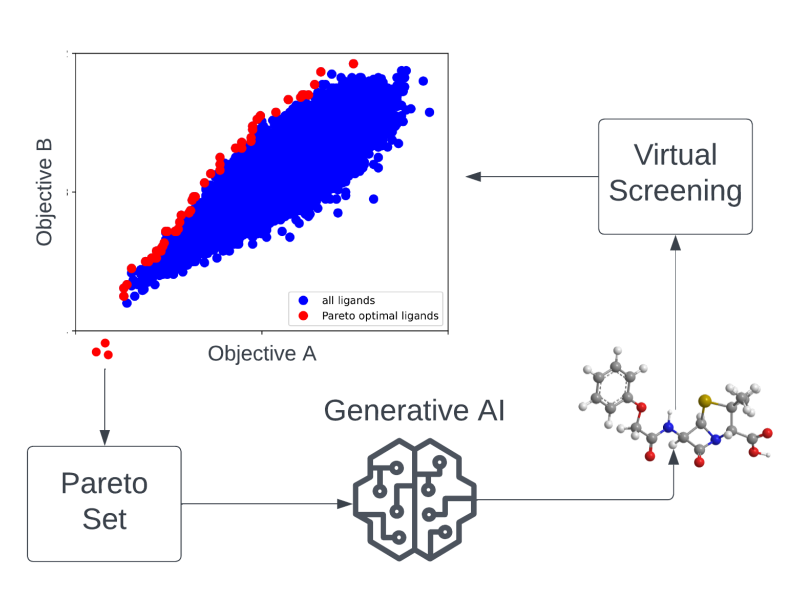
The search for novel drug candidates that, at the same time, act with high efficacy, comply with defined chemical properties, and also show low off-target effects can be perceived as a multi-objective optimization problem (MOOP) in chemical space. In the MATH+ Project "AA1-19 Drug Candidates as Pareto Optima in Chemical Space" AI-based generative models are used to identify novel drug-like molecules from chemical space beyond existing ligand libraries for therapeutic targets of unmet medical need. Due to the extremely large search space (approximately 10⁶⁰ drug-like molecules), efficient learning algorithms and high-throughput virtual screening methods are developed and applied in this project.
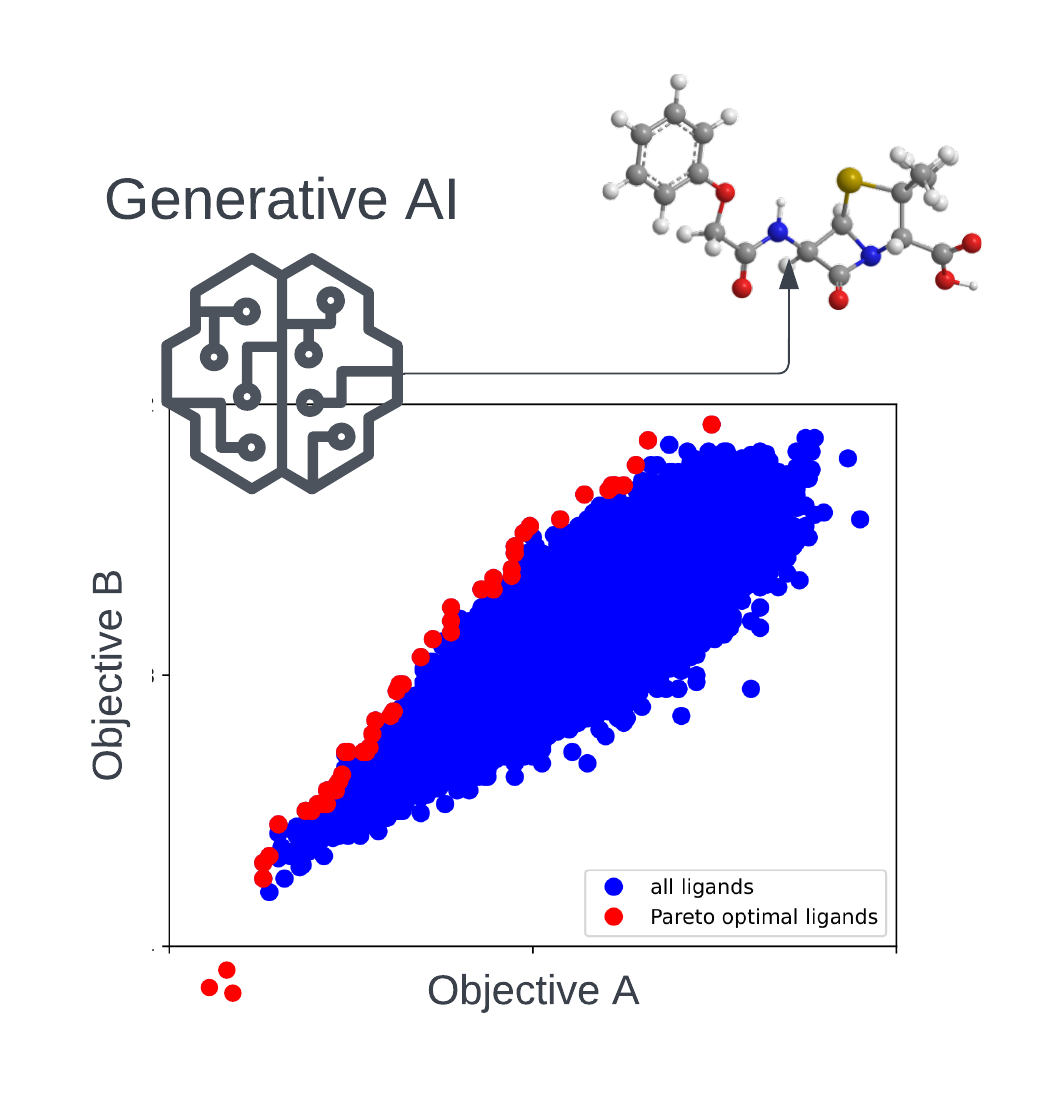 Generative AI for identifying Pareto-optimal drug candidates | In this project we are using generative AI to identify novel drug candidates that optimally combine multiple desired properties at the same time. |
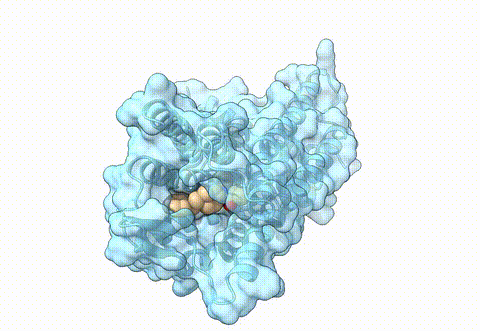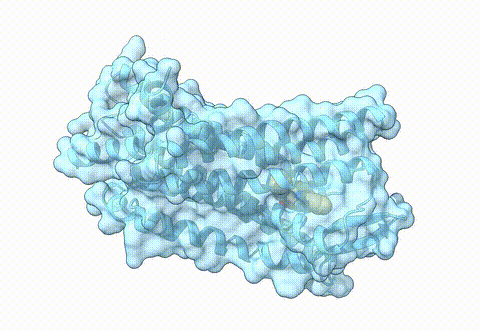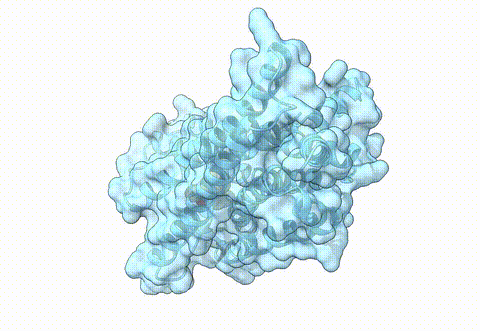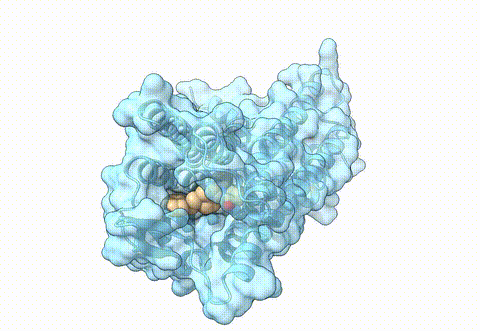 Structure of human µ-opioid receptor (PDB 8ef5) | Opioids are essential pharmaceuticals for pain relief, however, potential side effects are challenging. This class of molecules target the opioid receptors, for pain relief the main target represents the µ-opioid receptor (MOR). One approach to reduce side effects of MOR targeting drugs is to identify ligands that specifically target the receptor in an acidic, inflamed environment and bind with less affinity to the receptor in a neutral, non disease state, environment. In a differential virtual screening approach, we use high-throughput docking workflows together with generative AI to identify novel molecules with such properties. For experimental validation of pH-specific MOR agonists, we collaborate with the group of Dr. M. Özgür Çelik from the Anesthesiology Department at Charité Berlin. |
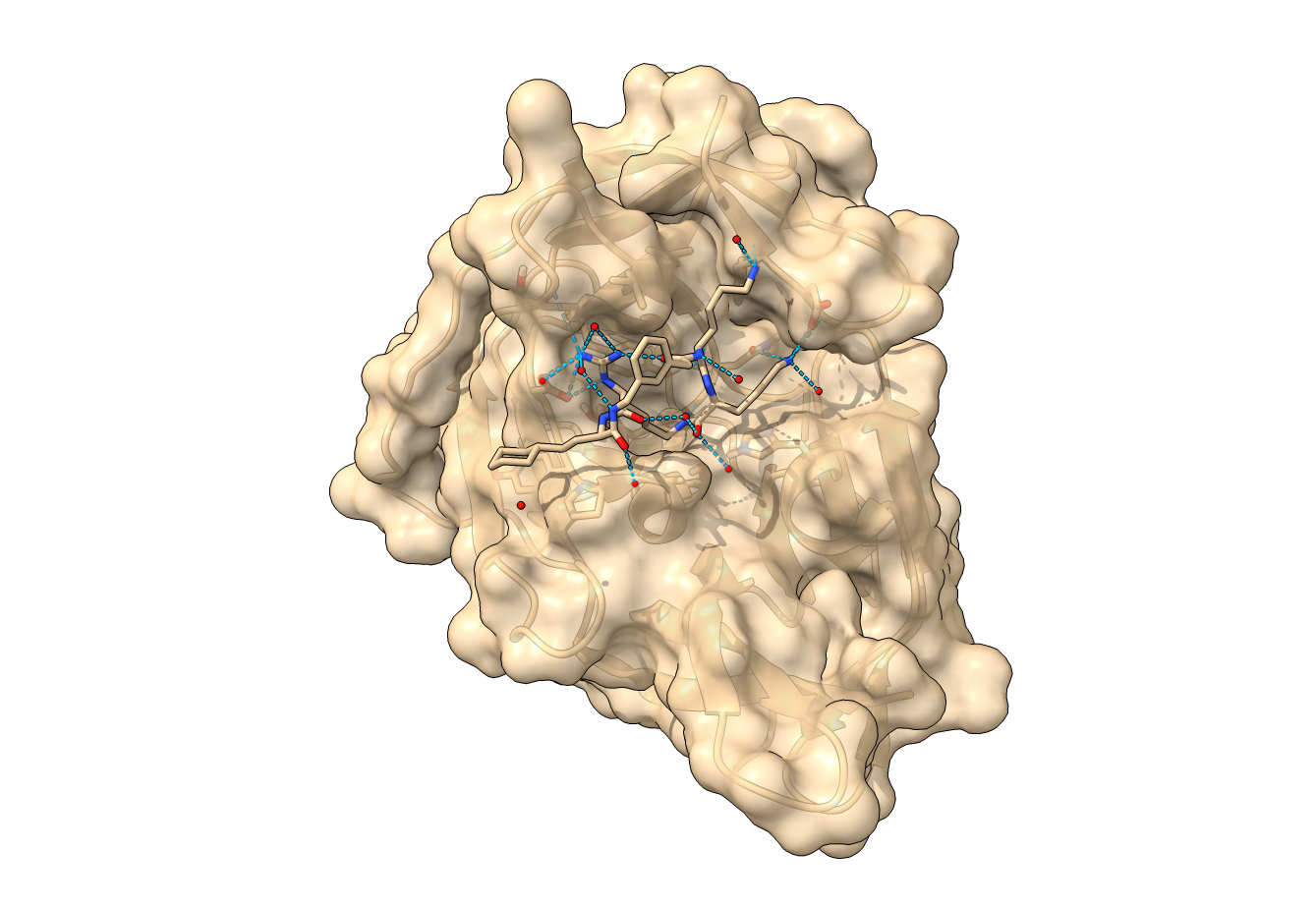 Structure of NS2B-NS3 flavivirus protease (PDB 7pgc) | Flaviviridae are a family of RNA viruses that can cause several different diseases in human. Among these are clinically challenging conditions including hepatitis, encephalitis, hemorrhagic fever and birth defects. To date, no effective therapeutic agent exists to prevent or ameliorate flavivirus-related disease. Therefore, we use our AI-guided virtual screening approach to identify novel drug candidates to target the NS2B-NS3 flavivirus protease. For experimental validation, we collaborate with the Medicinal Chemistry group of Prof. Jörg Rademann at the Institute of Pharmacy of Freie Universität (FU) Berlin. |