The causes of lower back pain (LBP) are still not fully understood. One essential part of a better understanding might be the association of LBP, spinal morphology, and kinematics. According to the Roussouly scheme [Roussouly et al., 2005] and some of its population-based computational extensions [F. Laouissat et. al, 2018], currently, spinal shapes are most commonly classified into four types, which only look at two-dimensional sagittal views. In other words, these classes do not consider the three-dimensional (3D) shape of the entire human spine. Although one previous study developed a 3D classification scheme, it does not correlate those classes with LBP and kinematics [J. Boisvert et. al, 2008].
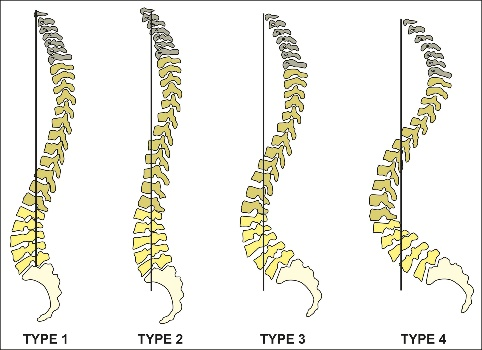
Figure 1: Roussouly scheme [Roussouly et al., 2005]
Our project is part of the DFG Research Unit 5177 “The Dynamics of the Spine: Mechanics, Morphology, and Motion towards a Comprehensive Diagnosis of Low Back Pain”, where we are investigating the spinal 3D morphology and motion in association with LBP based on cohort data.
For this purpose, we are developing a statistical 3D shape model (SSM) of the spine that will incorporate the variation in spinal morphology of the complete spinal shape and the individual vertebrae. Furthermore, we are building a fast articulation and deformation model of the spine that we can utilize to fit our SSM to any spinal shape and pose.
We will then individualize these shape models to static and dynamic measurements of subjects, which will help us to find correlations between spinal morphology and functional impairments or LBP. Another purpose of these individualized spines is to improve the accuracy of biomechanical analyses, which currently only utilize an average spinal shape.
Our main goal is to establish an extended morphological score for spinal diagnostics and further research.
Data Collection and Segmentation
For creating statistical shape models, we first collected a large pool of segmented medical spine imaging data: for instance the German National Cohort (GNC) [Bamberg et al., 2017], our study - the Berliner Rückenstudie, and the VerSe Challenge [Sekuboyina et al., 2021].
A special focus lies on the Berliner Rückenstudie, a study within our research collaboration. Our partners take extensive measurements of the human lumbar spine of up to 3000 subjects, and further process the data within the research unit. To generate ground truth data for training and testing automatic machine learning algorithms, our experts are segmenting individual vertebrae from tomographic CT and MR image data.
Limitations of MRI-Segmentations
Manual and automatic spinal structure segmentations from MRIs have several limitations (Figure 2). The low contrast between bones and surrounding soft tissue in MRIs leads to incomplete anatomical structures in the segmentations (structural sparsity). Furthermore, the high slice distance of the images leads to missing information between the slices (inter-slice sparsity). The image noise and the inhomogeneity of image intensity add to the difficulty of the segmentation process, especially for smaller vertebral structures.
These challenges make manual segmentation an expensive process that requires prior anatomical knowledge to interpolate invisible bony structures. Some existing algorithms try to solve this challenge by segmenting images in the CT domain, which is easier for bony structures. They then convert the CT images into MR images [Graf et. al, 2023], [Möller et. al, 2024]. This can lower the manual segmentation cost and improve the segmentation quality to some extent but they are still not as good as CT segmentation, especially w.r.t. the cervical spine. This could lead to inaccuracies in downstream tasks. Therefore segmentations from high-contrast and high-resolution CTs could be considered the gold standard for bone segmentations.
On the other hand, CT scanners expose the human body to higher radiation, which is why MR is the common choice in human studies.
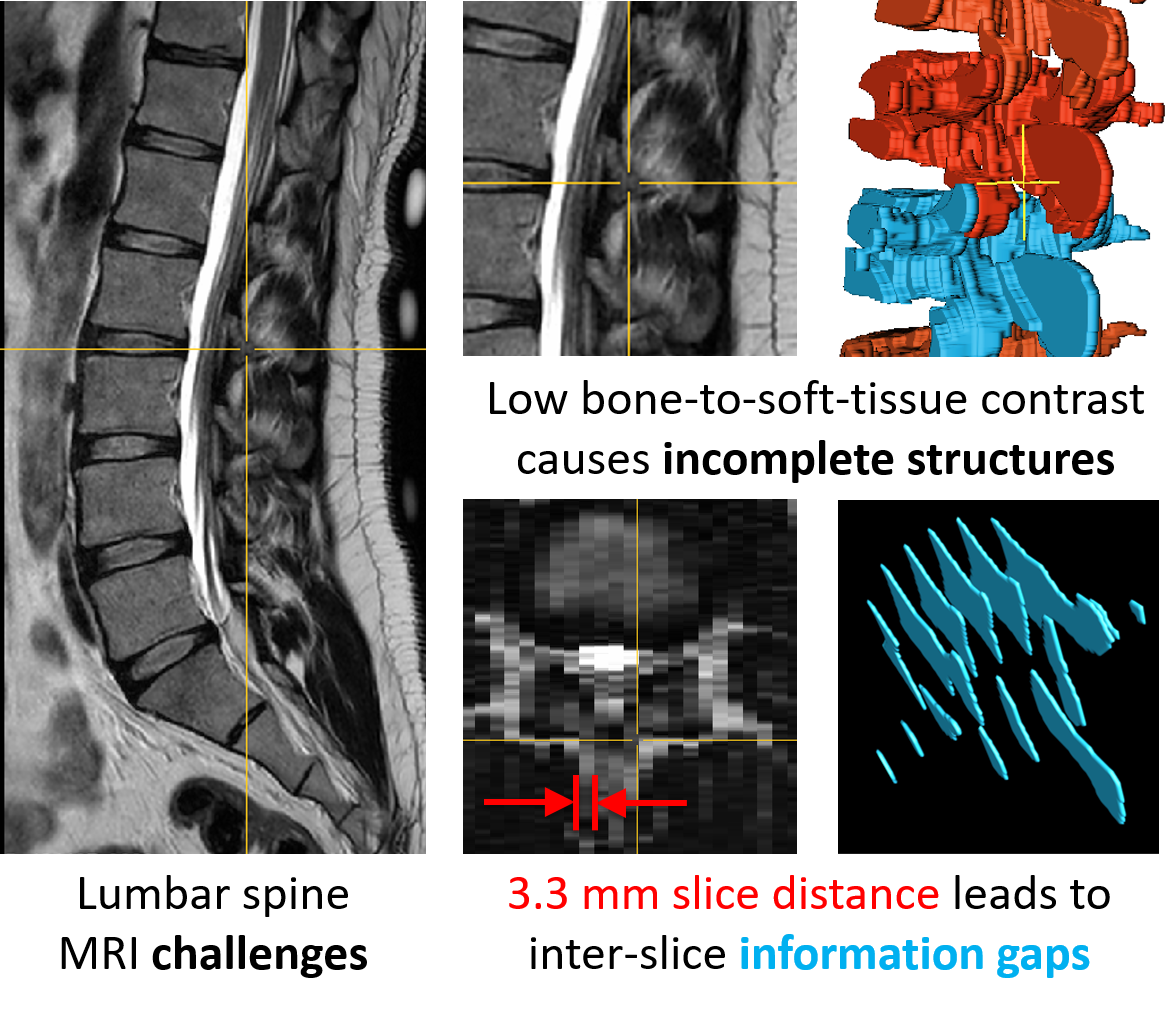
Figure 2: Challenges during MRI segmentation
Multi-class Statistical Shape Model
We aim to convert these low-resolution and incomplete segmentations into higher-resolution and complete CT-like segmentations. We are developing an SSM that learns from CT segmentations and transfers the knowledge to MR segmentations by interpolating the low-resolution and incomplete shapes to higher resolutions and in-paint the missing parts. A special focus lies on repairing individual vertebrae including the vertebral processes, arches, facets, and foramina. Furthermore, the SSM should also be used to predict a complete spine from only seeing a partial spine. Additionally, this SSM should provide a latent space representation to identify morphological clusters.
Shape Super-resolution
MRIs can have a slice distance higher than 3.3 mm, which leads to gaps in the segmentations. These inter-slice gaps need to be interpolated with plausible information.
The computer vision community has been researching 3D reconstruction techniques using neural networks. Recent work in our group has successfully reconstructed single vertebrae from sparse segmentation data [Amiranashvili et al., 2024] (Figure 3). These methods have been shown to work on single vertebrae without class information. However, our goal is to reconstruct the complete spine, where the semantics of each vertebra are represented (Figure 4).
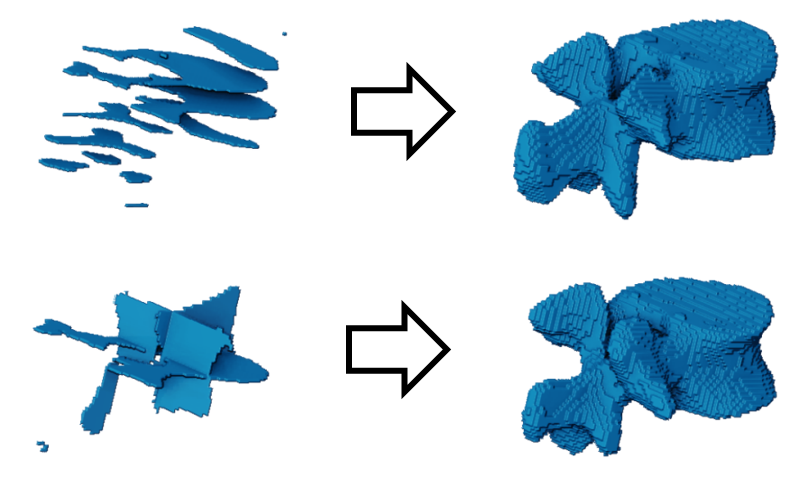
Figure 3: Reconstructing vertebrae from parallel (top) and orthogonal (bottom) slices [Amiranashvili et al., 2024].
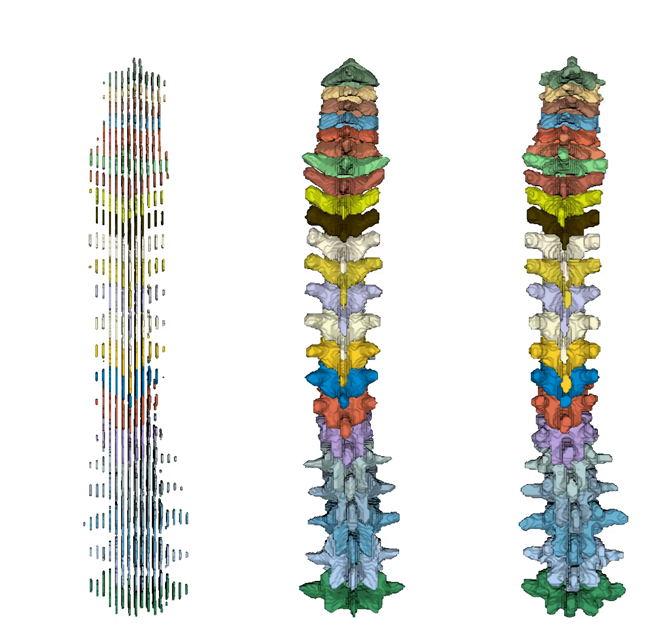
Figure 4: Super-resolution of the complete spine; from left to right: Low-resolution spine (3 mm slice distance), prediction, and ground truth (each 1 mm slice distance).
Shape Completion
Often medical images capture only a partial region of interest. However downstream tasks might need a complete shape. We are able to predict complete spines from only partial observations.
Figure 5: Completion and super-resolution of a partial spine; from left to right: Partial spine in lower-resolution (60% cropped and 3 mm slice distance), prediction, and ground truth (each 1 mm slice distance).
Articulation Models
The images of human spines are usually captured in a lying position but biomechanical analyses are conducted for upright spines. Furthermore, we only scan the lumbar spines in our research unit. If we want to predict a complete spine, we also need to adjust the pose. An articulation model can solve this by individualizing a spine to any pose (Figure 5).
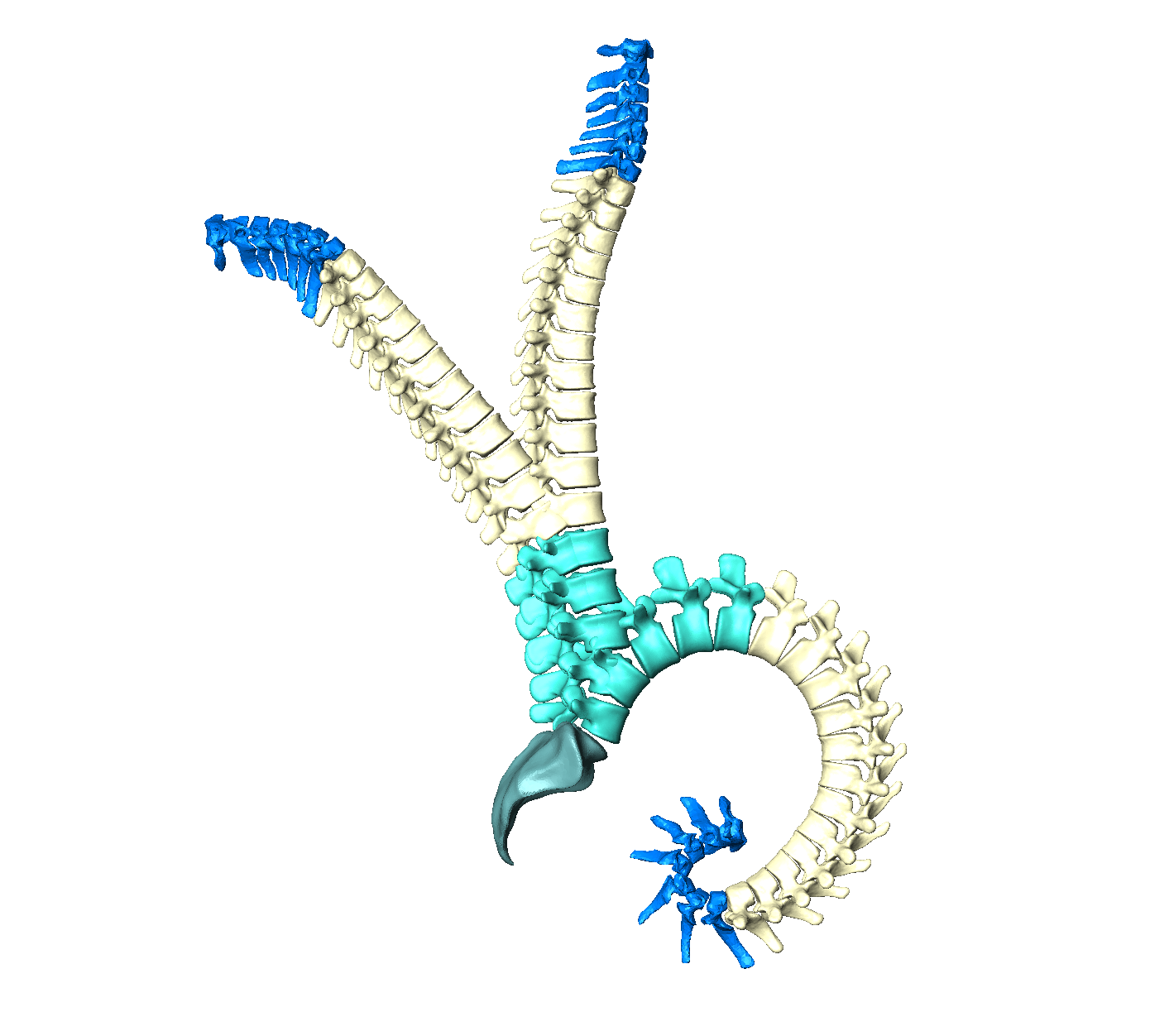
Figure 6: Articulation model of the human spine
Morphological Analysis of the Spine
We are analyzing variations of spinal shapes within a population as well as the relationship between spinal morphology and clinical patient parameters.
Principal Shape Variations
The first step of our spinal shape analysis is to detect the modes of variation. This may lead to a data-driven classification of spinal shapes, which provides a new perspective on spine morphology and help automatically diagnose spinal pathology.
Figure 7: Example mode of shape variation within a spinal population of 400 spines. The mean shape is displayed in the center and the multiples of the standard deviation is shown in positive and negative direction. At three times the standard deviation we reach the tails of our shape distribution, which can lead to artifacts if the distribution has less examples at this tail.
Associations between Morphology and Clinical Data
We aim to go deeper in our analysis by investigating the relationship between spinal morphology and clinical patient parameters.
References
| [Amiranashvili et al., 2024] | Learning continuous shape priors from sparse data with neural implicit functions, Medical Image Analysis. 2024 |
| [Möller et al., 2024] | Denoising diffusion-based MRI to CT image translation enables automated spinal segmentation. arxiv 2024 |
| [Graf et al., 2023] | SPINEPS – Automatic Whole Spine Segmentation of T2-weighted MR images using a Two-Phase Approach to Multi-class Semantic and Instance Segmentation. Europ. Radio. Exp. 2023 |
| [Sekuboyina et al., 2021] | VerSe: A Vertebrae Labelling and Segmentation Benchmark for Multi-detector CT Images. Medical Image Analysis 2021 |
| [F. Laouissat et al., 2018] | Classification of normal sagittal spine alignment: refounding the Roussouly classification. Europ. Spine Journ. 27(8): 2002–11, 2018. |
| [Bamberg et al., 2017] | Whole-Body MR Imaging in the German National Cohort: Rationale, Design, and Technical Background. Radiology 277(1): 206 – 20, 2015. |
| [J. Boisvert et al., 2008] | Principal Deformations Modes of Articulated Models for the Analysis of 3D Spine Deformities. Letters on Comp. Vis. & Imag. Anal. 7(4):13–31, 2008. |
| [Roussouly et al., 2005] | Classification of the normal variation in the sagittal alignment of the human lumbar spine and pelvis in the standing position. Spine 2005 |