Fully automatic segmentation of arbitrary anatomical structures from 3D medical image data is a challenging, yet unsolved problem. Though fully automatic segmentation is essential for further clinical analysis, complexity of anatomical structures across population makes a generalized segmentation scheme extremely challenging. Moreover, specific challenges of different imaging modalities have so far hindered the possibility of a general purpose fully automatic 3D segmentation framework. Statistical 3D shape models have proven to be valuable shape priors that are to be deformed within their range of normal variation in shape to match the respective image information. Within the project, we are aiming to combine Machine Learning along with the statistical shape priors for getting a step closer to a general 3D image segmentation approach. In particular, Machine Learning techniques for image matching based on intensity will be developed in order to improve both the model building as well as the segmentation process.
Image-based Cost Functions
Principal Component Analysis (PCA) on local intensity profiles has not proven to beneficially act as a robust cost function. Random Forest Regression Voting (RFRV), though a powerful method for 2D image data, turned out to be impractical for 3D data, due to huge memory consumption and computational time. Dictionary Learning (DL) does not require any heuristics and is general enough to be applied across anatomies and modalities. DL operations are matrix operations, thus being efficiently evaluated.
Joint Dictionary Learning
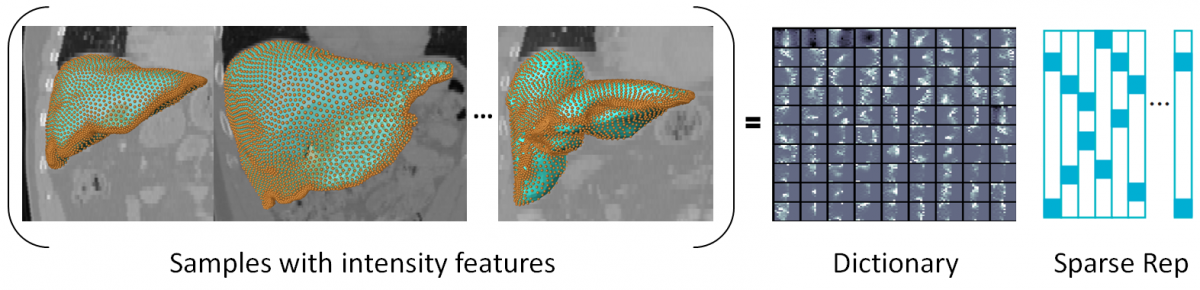
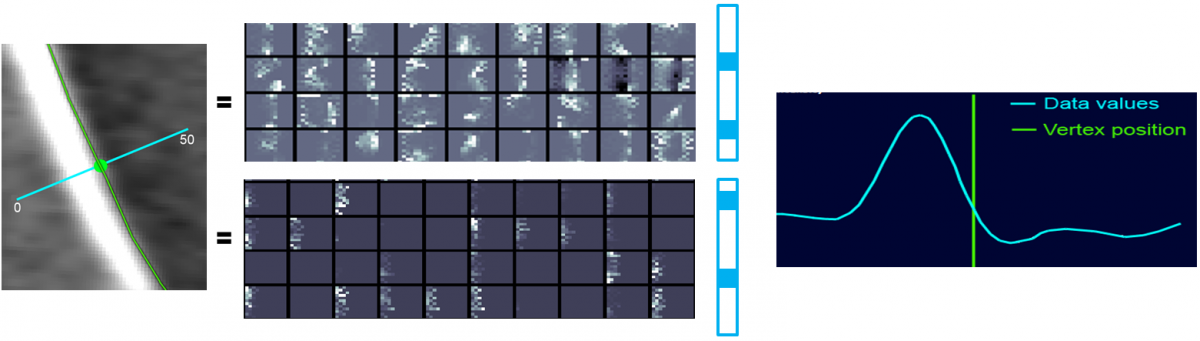
Given 3D image data and accordingly segmented anatomical structures of interest, rotational invariant histograms of oriented gradients (HoG) are sampled at the structures’ boundaries. These feature samples are used as input for learning a dictionary.
A second dictionary is learnt for background image information. A combined dictionary of foreground and background features has been established, acting as a cost function for image segmentation.
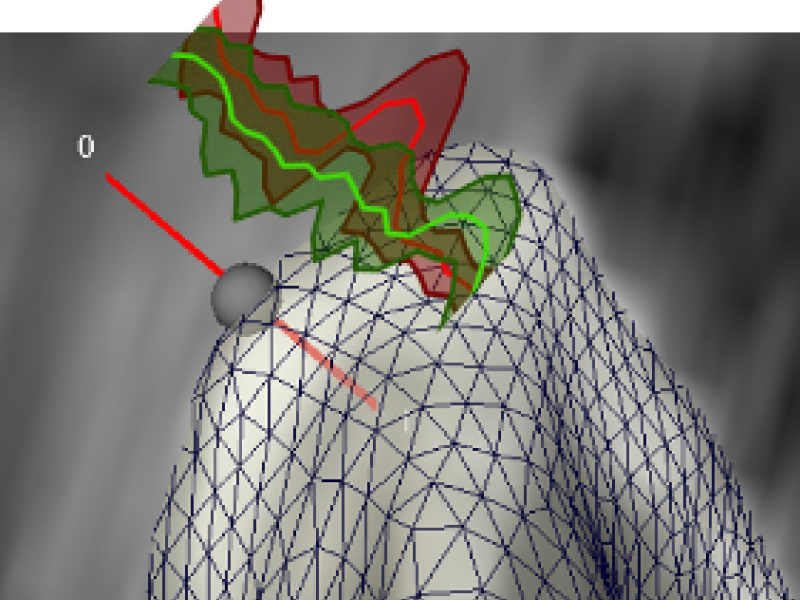
Cost Function for a test patch: Sum of residuals from representations by the two dictionaries.
Clustered Appearance Models
Local appearance patterns of anatomical structures in medical image data play an important role in determining the location, yet the variation of these patterns across localities are challenging for traditional appearance models. Dictionary Learning, even though widely used as an appearance model, lacks a systematic approach for comparison between dictionaries for subsequent clustering. A dictionary comparison framework is proposed for generating clustered appearance models useful for fully automatic segmentation.
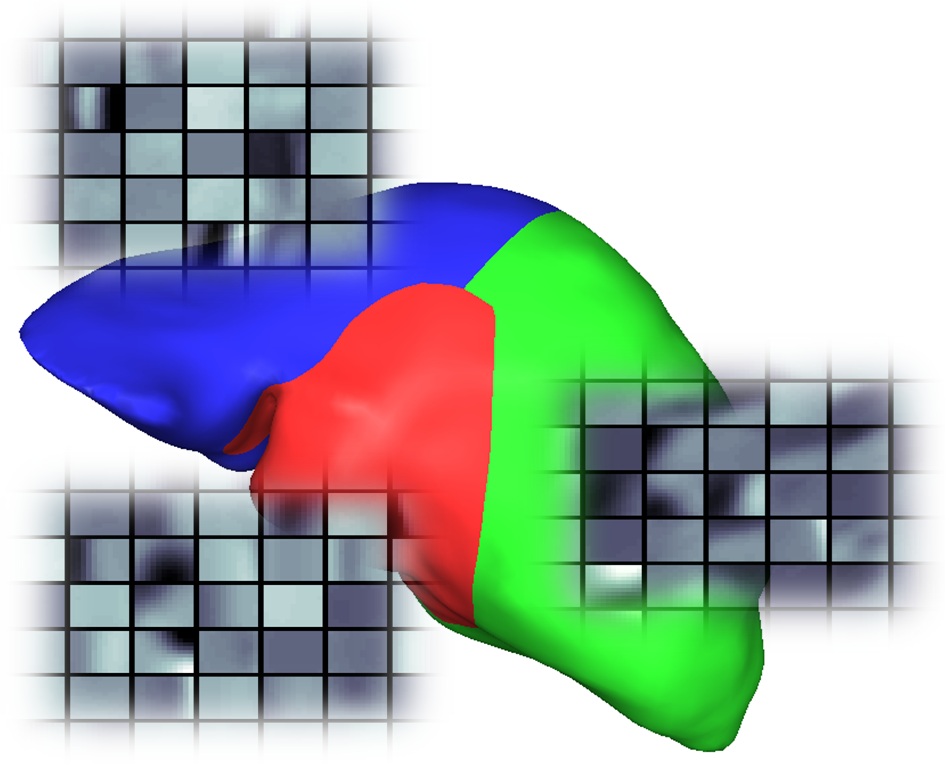
We consider available dictionary comparison metrics and propose two new metrics for comparison. Three novel meta comparison schemes are also developed for systematic comparison of the metrics in appearance dictionary context. Furthermore, a novel non-overlapping dictionary clustering approach is developed for clustered appearance modeling on anatomical structures.
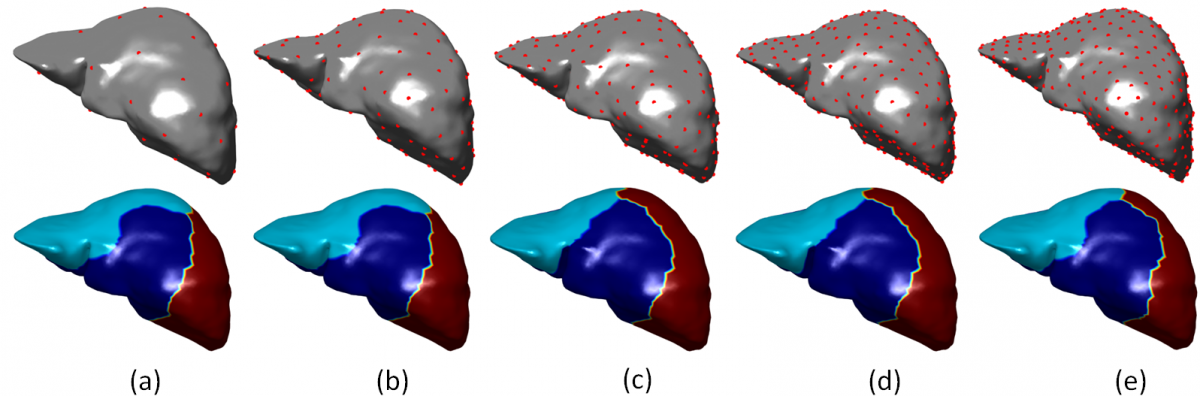
Initial Seeds (upper row) and the resulting clusters (bottom row) by our dictionary clustering algorithm. Notice the similarity of clusters for di fferent number of initial seeds: (a) 50, (b) 150, (c) 250, (d) 350 and (e) 450.